Congenital disorders
Exploring the molecular and clinical spectrum of COVID-19-related acute necrotizing encephalopathy in three pediatric cases
Abstract
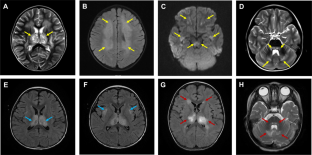
Acute necrotizing encephalopathy (ANE) is a rare disease that predominantly affects children and is associated with a high mortality rate. Here we report three cases of COVID-19-related ANE in children, with the mutation detection in two genes associated with mitochondrial dysfunction. The cases exhibited common ANE symptoms, such as fever, impaired consciousness, positive pathological reflex, increased cerebrospinal fluid protein, and multifocal and symmetric brain lesions identified through MRI. Using genotype-phenotype correlation analysis in trio-whole exome sequencing (WES), four potential pathogenic variants were identified in two genes associated with mitochondrial function (RANBP2 and MCCC2). Notably, MCCC2 was identified as being potentially associated with COVID-19-related ANE for the first time, and two of the four variants had not been previously reported. Our findings expand the clinical and mutation spectrum of COVID-19-related ANE in pediatric cases. The finding of these three new cases in our study further supports the previous hypothesis about the role of mitochondrial homeostatic imbalance in COVID-19-related ANE. It is essential to use genetic testing to identify this subset of patients with compromised mitochondrial function in order to improve patient management and prognosis.
This is a preview of subscription content, access via your institution
Access options
style{display:none!important}.LiveAreaSection-193358632 *{align-content:stretch;align-items:stretch;align-self:auto;animation-delay:0s;animation-direction:normal;animation-duration:0s;animation-fill-mode:none;animation-iteration-count:1;animation-name:none;animation-play-state:running;animation-timing-function:ease;azimuth:center;backface-visibility:visible;background-attachment:scroll;background-blend-mode:normal;background-clip:borderBox;background-color:transparent;background-image:none;background-origin:paddingBox;background-position:0 0;background-repeat:repeat;background-size:auto auto;block-size:auto;border-block-end-color:currentcolor;border-block-end-style:none;border-block-end-width:medium;border-block-start-color:currentcolor;border-block-start-style:none;border-block-start-width:medium;border-bottom-color:currentcolor;border-bottom-left-radius:0;border-bottom-right-radius:0;border-bottom-style:none;border-bottom-width:medium;border-collapse:separate;border-image-outset:0s;border-image-repeat:stretch;border-image-slice:100%;border-image-source:none;border-image-width:1;border-inline-end-color:currentcolor;border-inline-end-style:none;border-inline-end-width:medium;border-inline-start-color:currentcolor;border-inline-start-style:none;border-inline-start-width:medium;border-left-color:currentcolor;border-left-style:none;border-left-width:medium;border-right-color:currentcolor;border-right-style:none;border-right-width:medium;border-spacing:0;border-top-color:currentcolor;border-top-left-radius:0;border-top-right-radius:0;border-top-style:none;border-top-width:medium;bottom:auto;box-decoration-break:slice;box-shadow:none;box-sizing:border-box;break-after:auto;break-before:auto;break-inside:auto;caption-side:top;caret-color:auto;clear:none;clip:auto;clip-path:none;color:initial;column-count:auto;column-fill:balance;column-gap:normal;column-rule-color:currentcolor;column-rule-style:none;column-rule-width:medium;column-span:none;column-width:auto;content:normal;counter-increment:none;counter-reset:none;cursor:auto;display:inline;empty-cells:show;filter:none;flex-basis:auto;flex-direction:row;flex-grow:0;flex-shrink:1;flex-wrap:nowrap;float:none;font-family:initial;font-feature-settings:normal;font-kerning:auto;font-language-override:normal;font-size:medium;font-size-adjust:none;font-stretch:normal;font-style:normal;font-synthesis:weight style;font-variant:normal;font-variant-alternates:normal;font-variant-caps:normal;font-variant-east-asian:normal;font-variant-ligatures:normal;font-variant-numeric:normal;font-variant-position:normal;font-weight:400;grid-auto-columns:auto;grid-auto-flow:row;grid-auto-rows:auto;grid-column-end:auto;grid-column-gap:0;grid-column-start:auto;grid-row-end:auto;grid-row-gap:0;grid-row-start:auto;grid-template-areas:none;grid-template-columns:none;grid-template-rows:none;height:auto;hyphens:manual;image-orientation:0deg;image-rendering:auto;image-resolution:1dppx;ime-mode:auto;inline-size:auto;isolation:auto;justify-content:flexStart;left:auto;letter-spacing:normal;line-break:auto;line-height:normal;list-style-image:none;list-style-position:outside;list-style-type:disc;margin-block-end:0;margin-block-start:0;margin-bottom:0;margin-inline-end:0;margin-inline-start:0;margin-left:0;margin-right:0;margin-top:0;mask-clip:borderBox;mask-composite:add;mask-image:none;mask-mode:matchSource;mask-origin:borderBox;mask-position:0 0;mask-repeat:repeat;mask-size:auto;mask-type:luminance;max-height:none;max-width:none;min-block-size:0;min-height:0;min-inline-size:0;min-width:0;mix-blend-mode:normal;object-fit:fill;object-position:50% 50%;offset-block-end:auto;offset-block-start:auto;offset-inline-end:auto;offset-inline-start:auto;opacity:1;order:0;orphans:2;outline-color:initial;outline-offset:0;outline-style:none;outline-width:medium;overflow:visible;overflow-wrap:normal;overflow-x:visible;overflow-y:visible;padding-block-end:0;padding-block-start:0;padding-bottom:0;padding-inline-end:0;padding-inline-start:0;padding-left:0;padding-right:0;padding-top:0;page-break-after:auto;page-break-before:auto;page-break-inside:auto;perspective:none;perspective-origin:50% 50%;pointer-events:auto;position:static;quotes:initial;resize:none;right:auto;ruby-align:spaceAround;ruby-merge:separate;ruby-position:over;scroll-behavior:auto;scroll-snap-coordinate:none;scroll-snap-destination:0 0;scroll-snap-points-x:none;scroll-snap-points-y:none;scroll-snap-type:none;shape-image-threshold:0;shape-margin:0;shape-outside:none;tab-size:8;table-layout:auto;text-align:initial;text-align-last:auto;text-combine-upright:none;text-decoration-color:currentcolor;text-decoration-line:none;text-decoration-style:solid;text-emphasis-color:currentcolor;text-emphasis-position:over right;text-emphasis-style:none;text-indent:0;text-justify:auto;text-orientation:mixed;text-overflow:clip;text-rendering:auto;text-shadow:none;text-transform:none;text-underline-position:auto;top:auto;touch-action:auto;transform:none;transform-box:borderBox;transform-origin:50% 50%0;transform-style:flat;transition-delay:0s;transition-duration:0s;transition-property:all;transition-timing-function:ease;vertical-align:baseline;visibility:visible;white-space:normal;widows:2;width:auto;will-change:auto;word-break:normal;word-spacing:normal;word-wrap:normal;writing-mode:horizontalTb;z-index:auto;-webkit-appearance:none;-moz-appearance:none;-ms-appearance:none;appearance:none;margin:0}.LiveAreaSection-193358632{width:100%}.LiveAreaSection-193358632 .login-option-buybox{display:block;width:100%;font-size:17px;line-height:30px;color:#222;padding-top:30px;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-access-options{display:block;font-weight:700;font-size:17px;line-height:30px;color:#222;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-login>li:not(:first-child)::before{transform:translateY(-50%);content:””;height:1rem;position:absolute;top:50%;left:0;border-left:2px solid #999}.LiveAreaSection-193358632 .additional-login>li:not(:first-child){padding-left:10px}.LiveAreaSection-193358632 .additional-login>li{display:inline-block;position:relative;vertical-align:middle;padding-right:10px}.BuyBoxSection-683559780{display:flex;flex-wrap:wrap;flex:1;flex-direction:row-reverse;margin:-30px -15px 0}.BuyBoxSection-683559780 .box-inner{width:100%;height:100%}.BuyBoxSection-683559780 .readcube-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:1;flex-basis:255px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:300px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox-nature-plus{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:100%;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .title-readcube,.BuyBoxSection-683559780 .title-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .title-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .asia-link{color:#069;cursor:pointer;text-decoration:none;font-size:1.05em;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:1.05em6}.BuyBoxSection-683559780 .access-readcube{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;opacity:.8px;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .price-buybox{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;padding-top:30px;text-align:center}.BuyBoxSection-683559780 .price-buybox-to{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center}.BuyBoxSection-683559780 .price-info-text{font-size:16px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-value{font-size:30px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-per-period{font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-from{font-size:14px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .issue-buybox{display:block;font-size:13px;text-align:center;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:19px}.BuyBoxSection-683559780 .no-price-buybox{display:block;font-size:13px;line-height:18px;text-align:center;padding-right:10%;padding-left:10%;padding-bottom:20px;padding-top:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .vat-buybox{display:block;margin-top:5px;margin-right:20%;margin-left:20%;font-size:11px;color:#222;padding-top:10px;padding-bottom:15px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:17px}.BuyBoxSection-683559780 .tax-buybox{display:block;width:100%;color:#222;padding:20px 16px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:NaNpx}.BuyBoxSection-683559780 .button-container{display:flex;padding-right:20px;padding-left:20px;justify-content:center}.BuyBoxSection-683559780 .button-container>*{flex:1px}.BuyBoxSection-683559780 .button-container>a:hover,.Button-1078489254:hover,.Button-2496381730:hover{text-decoration:none}.BuyBoxSection-683559780 .readcube-button{background:#fff;margin-top:30px}.BuyBoxSection-683559780 .button-asia{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;margin-top:75px}.BuyBoxSection-683559780 .button-label-asia,.ButtonLabel-3296148077,.ButtonLabel-1651148777{display:block;color:#fff;font-size:17px;line-height:20px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center;text-decoration:none;cursor:pointer}.Button-1078489254,.Button-2496381730{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;max-width:320px;margin-top:10px}.Button-1078489254 .readcube-label,.Button-2496381730 .readcube-label{color:#069}
/* style specs end */
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout



References
-
Messiah SE, Xie L, Mathew MS, Delclos GL, Kohl HW 3rd, Kahn JS. Results of COVID-19 surveillance in a large United States pediatric healthcare system over one year. Child (Basel). 2021;8:752.
-
Cloete J, Kruger A, Masha M, du Plessis NM, Mawela D, Tshukudu M, et al. Paediatric hospitalisations due to COVID-19 during the first SARS-CoV-2 omicron (B.1.1.529) variant wave in South Africa: a multicentre observational study. Lancet Child Adolesc Health. 2022;6:294–302.
Google Scholar
-
Ross CE, Burns JP, Grossestreuer AV, Bhattarai P, McKiernan CA, Franks JD, et al. Trends in disease severity among critically Ill children with severe acute respiratory syndrome coronavirus 2: a retrospective multicenter cohort study in the United States. Pediatr Crit Care Med. 2023;24:25–33.
Google Scholar
-
Chi H, Chang L, Chao YC, Lin DS, Yang HW, Fang LC, et al. Pathogenesis and preventive tactics of immune-mediated non-pulmonary COVID-19 in children and beyond. Int J Mol Sci. 2022;23:14157.
Google Scholar
-
Ray STJ, Abdel-Mannan O, Sa M, Fuller C, Wood GK, Pysden K, et al. Neurological manifestations of SARS-CoV-2 infection in hospitalised children and adolescents in the UK: a prospective national cohort study. Lancet Child Adolesc Health. 2021;5:631–41.
Google Scholar
-
Islam MA, Cavestro C, Alam SS, Kundu S, Kamal MA, Reza F. Encephalitis in patients with COVID-19: a systematic evidence-based analysis. Cells. 2022;11:2575.
Google Scholar
-
Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–60.
Google Scholar
-
Pilotto A, Masciocchi S, Volonghi I, De Giuli V, Caprioli F, Mariotto S, et al. Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) encephalitis is a cytokine release syndrome: evidences from cerebrospinal fluid analyses. Clin Infect Dis. 2021;73:e3019–e26.
Google Scholar
-
Lee MH, Perl DP, Steiner J, Pasternack N, Li W, Maric D, et al. Neurovascular injury with complement activation and inflammation in COVID-19. Brain. 2022;145:2555–68.
Google Scholar
-
Mizuguchi M. Acute necrotizing encephalopathy of childhood: a novel form of acute encephalopathy prevalent in Japan and Taiwan. Brain Dev. 1997;19:81–92.
Google Scholar
-
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–9.
Google Scholar
-
Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–80.
Google Scholar
-
Strom SP, Hossain WA, Grigorian M, Li M, Fierro J, Scaringe W, et al. A streamlined approach to prader-willi and angelman syndrome molecular diagnostics. Front Genet. 2021;12:608889.
Google Scholar
-
Kohler S, Gargano M, Matentzoglu N, Carmody LC, Lewis-Smith D, Vasilevsky NA, et al. The human phenotype ontology in 2021. Nucleic Acids Res. 2021;49:D1207–D17.
Google Scholar
-
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–24.
Google Scholar
-
Capalbo A, Valero RA, Jimenez-Almazan J, Pardo PM, Fabiani M, Jimenez D, et al. Optimizing clinical exome design and parallel gene-testing for recessive genetic conditions in preconception carrier screening: translational research genomic data from 14,125 exomes. PLoS Genet. 2019;15:e1008409.
Google Scholar
-
Molina-Ramirez LP, Kyle C, Ellingford JM, Wright R, Taylor A, Bhaskar SS, et al. Personalised virtual gene panels reduce interpretation workload and maintain diagnostic rates of proband-only clinical exome sequencing for rare disorders. J Med Genet. 2022;59:393–98.
Google Scholar
-
Neilson DE, Adams MD, Orr CM, Schelling DK, Eiben RM, Kerr DS, et al. Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am J Hum Genet. 2009;84:44–51.
Google Scholar
-
Grunert SC, Stucki M, Morscher RJ, Suormala T, Burer C, Burda P, et al. 3-methylcrotonyl-CoA carboxylase deficiency: clinical, biochemical, enzymatic and molecular studies in 88 individuals. Orphanet J Rare Dis. 2012;7:31.
Google Scholar
-
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Google Scholar
-
Jiang J, Wang YE, Palazzo AF, Shen Q. Roles of nucleoporin RanBP2/Nup358 in acute necrotizing encephalopathy type 1 (ANE1) and viral infection. Int J Mol Sci. 2022;23:3548.
Google Scholar
-
Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–15.
Google Scholar
-
Liu Y, Chang X, Glessner J, Qu H, Tian L, Li D, et al. Association of rare recurrent copy number variants with congenital heart defects based on next-generation sequencing data from family trios. Front Genet. 2019;10:819.
Google Scholar
-
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Google Scholar
-
Pejaver V, Byrne AB, Feng BJ, Pagel KA, Mooney SD, Karchin R, et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am J Hum Genet. 2022;109:2163–77.
Google Scholar
-
Zamani R, Pouremamali R, Rezaei N. Central neuroinflammation in Covid-19: a systematic review of 182 cases with encephalitis, acute disseminated encephalomyelitis, and necrotizing encephalopathies. Rev Neurosci. 2022;33:397–412.
Google Scholar
-
Wertheimer GSO, Brandao MB, Reis F. COVID-19-related acute necrotizing encephalopathy with new spectroscopy features. Rev Soc Bras Med Trop. 2022;55:e02752022.
Google Scholar
-
Khan M, Bhattarai S, Boyce TG, Hayek RA, Zhadanov SI, Hooper EE, et al. Acute necrotizing encephalopathy associated with coronavirus disease 2019 in an infant. J Pediatr. 2022;247:160–62.
Google Scholar
-
Lazarte-Rantes C, Guevara-Castanon J, Romero L, Guillen-Pinto D. Acute necrotizing encephalopathy associated with SARS-CoV-2 exposure in a pediatric patient. Cureus. 2021;13:e15018.
Google Scholar
-
Forest C, Laudisi M, Malaventura C, Tugnoli V, Pellino G, Marangoni E, et al. Pediatric recurrent acute necrotizing encephalomyelitis, RANBP2 genotype and Sars-CoV-2 infection: diagnosis, pathogenesis and targeted treatments from a case study. Eur J Paediatr Neurol. 2023;42:117–21.
Google Scholar
-
Palazzo AF, Joseph J, Lim M, Thakur KT. Workshop on RanBP2/Nup358 and acute necrotizing encephalopathy. Nucleus. 2022;13:154–69.
Google Scholar
-
Anand G, Visagan R, Chandratre S, Segal S, Nemeth AH, Squier W, et al. H1N1 triggered recurrent acute necrotizing encephalopathy in a family with a T653I mutation in the RANBP2 gene. Pediatr Infect Dis J. 2015;34:318–20.
Google Scholar
-
Hartley M, Sinha A, Kumar A, Aliu E, Mainali G, Paudel S. Acute necrotizing encephalopathy: 2 case reports on RANBP2 mutation. Child Neurol Open. 2021;8:2329048X211030751.
Google Scholar
-
Sell K, Storch K, Hahn G, Lee-Kirsch MA, Ramantani G, Jackson S, et al. Variable clinical course in acute necrotizing encephalopathy and identification of a novel RANBP2 mutation. Brain Dev. 2016;38:777–80.
Google Scholar
-
Stranneheim H, Lagerstedt-Robinson K, Magnusson M, Kvarnung M, Nilsson D, Lesko N, et al. Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med. 2021;13:40.
Google Scholar
-
Levine JM, Ahsan N, Ho E, Santoro JD. Genetic acute necrotizing encephalopathy associated with RANBP2: clinical and therapeutic implications in pediatrics. Mult Scler Relat Disord. 2020;43:102194.
Google Scholar
-
Cecchini R, Cecchini AL. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med Hypotheses. 2020;143:110102.
Google Scholar
-
Galan F, Nordli DR, Yazdani M, Klein J. A favorable treatment outcome in RANBP2 and influenza associated acute necrotizing encephalitis. Child Neurol Open. 2022;9:2329048X221143689.
Google Scholar
-
Gallardo ME, Desviat LR, Rodriguez JM, Esparza-Gordillo J, Perez-Cerda C, Perez B, et al. The molecular basis of 3-methylcrotonylglycinuria, a disorder of leucine catabolism. Am J Hum Genet. 2001;68:334–46.
Google Scholar
-
Baumgartner MR, Almashanu S, Suormala T, Obie C, Cole RN, Packman S, et al. The molecular basis of human 3-methylcrotonyl-CoA carboxylase deficiency. J Clin Invest. 2001;107:495–504.
Google Scholar
-
Sahin S, Yildirim M, Bektas O, Surucu Kara I, Ceylan AC, Teber S. Intracranial calcification associated with 3-methylcrotonyl-CoA carboxylase deficiency. Mol Syndromol. 2021;12:393–98.
Google Scholar
-
Elesela S, Lukacs NW. Role of mitochondria in viral infections. Life (Basel). 2021;11:232.
Google Scholar
-
Shibata A, Kasai M, Hoshino A, Tanaka T, Mizuguchi M. RANBP2 mutation causing autosomal dominant acute necrotizing encephalopathy attenuates its interaction with COX11. Neurosci Lett. 2021;763:136173.
Google Scholar
-
Al-Anezi A, Sotirova-Koulli V, Shalaby O, Ibrahim A, Abdulmotagalli N, Youssef R, et al. Biotin-thiamine responsive basal ganglia disease in the era of COVID-19 outbreak diagnosis not to be missed: a case report. Brain Dev. 2022;44:303–07.
Google Scholar
-
Thomsen JA, Lund AM, Olesen JH, Mohr M, Rasmussen J. Is L-Carnitine supplementation beneficial in 3-methylcrotonyl-CoA carboxylase deficiency? JIMD Rep. 2015;21:79–88.
Google Scholar
Acknowledgements
We would like to sincerely thank Professor Taosheng Huang for his invaluable advice and guidance in the writing and revision of the manuscript. We would also like to express our gratitude to all those who have provided assistance and support for our article.
Funding
This research was supported by the Natural Science Foundation of Fujian Province (Grant no. 2022J011097).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text 1
Supplementary Text 2
Supplementary Text 3
Supplementary Table S1
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and Permissions
About this article
Cite this article
Wu, D., Zheng, Y., Li, Y. et al. Exploring the molecular and clinical spectrum of COVID-19-related acute necrotizing encephalopathy in three pediatric cases.
J Hum Genet (2023). https://doi.org/10.1038/s10038-023-01171-z
-
Received: 03 March 2023
-
Revised: 04 June 2023
-
Accepted: 04 June 2023
-
Published: 25 July 2023
-
DOI: https://doi.org/10.1038/s10038-023-01171-z