Infection
Antibiotic-induced collateral damage to the microbiota and associated infections
Abstract
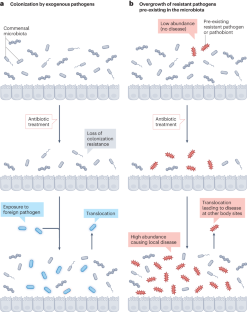
Antibiotics have transformed medicine, saving millions of lives since they were first used to treat a bacterial infection. However, antibiotics administered to target a specific pathogen can also cause collateral damage to the patient’s resident microbial population. These drugs can suppress the growth of commensal species which provide protection against colonization by foreign pathogens, leading to an increased risk of subsequent infection. At the same time, a patient’s microbiota can harbour potential pathogens and, hence, be a source of infection. Antibiotic-induced selection pressure can cause overgrowth of resistant pathogens pre-existing in the patient’s microbiota, leading to hard-to-treat superinfections. In this Review, we explore our current understanding of how antibiotic therapy can facilitate subsequent infections due to both loss of colonization resistance and overgrowth of resistant microorganisms, and how these processes are often interlinked. We discuss both well-known and currently overlooked examples of antibiotic-associated infections at various body sites from various pathogens. Finally, we describe ongoing and new strategies to overcome the collateral damage caused by antibiotics and to limit the risk of antibiotic-associated infections.
This is a preview of subscription content, access via your institution
Access options
style{display:none!important}.LiveAreaSection-193358632 *{align-content:stretch;align-items:stretch;align-self:auto;animation-delay:0s;animation-direction:normal;animation-duration:0s;animation-fill-mode:none;animation-iteration-count:1;animation-name:none;animation-play-state:running;animation-timing-function:ease;azimuth:center;backface-visibility:visible;background-attachment:scroll;background-blend-mode:normal;background-clip:borderBox;background-color:transparent;background-image:none;background-origin:paddingBox;background-position:0 0;background-repeat:repeat;background-size:auto auto;block-size:auto;border-block-end-color:currentcolor;border-block-end-style:none;border-block-end-width:medium;border-block-start-color:currentcolor;border-block-start-style:none;border-block-start-width:medium;border-bottom-color:currentcolor;border-bottom-left-radius:0;border-bottom-right-radius:0;border-bottom-style:none;border-bottom-width:medium;border-collapse:separate;border-image-outset:0s;border-image-repeat:stretch;border-image-slice:100%;border-image-source:none;border-image-width:1;border-inline-end-color:currentcolor;border-inline-end-style:none;border-inline-end-width:medium;border-inline-start-color:currentcolor;border-inline-start-style:none;border-inline-start-width:medium;border-left-color:currentcolor;border-left-style:none;border-left-width:medium;border-right-color:currentcolor;border-right-style:none;border-right-width:medium;border-spacing:0;border-top-color:currentcolor;border-top-left-radius:0;border-top-right-radius:0;border-top-style:none;border-top-width:medium;bottom:auto;box-decoration-break:slice;box-shadow:none;box-sizing:border-box;break-after:auto;break-before:auto;break-inside:auto;caption-side:top;caret-color:auto;clear:none;clip:auto;clip-path:none;color:initial;column-count:auto;column-fill:balance;column-gap:normal;column-rule-color:currentcolor;column-rule-style:none;column-rule-width:medium;column-span:none;column-width:auto;content:normal;counter-increment:none;counter-reset:none;cursor:auto;display:inline;empty-cells:show;filter:none;flex-basis:auto;flex-direction:row;flex-grow:0;flex-shrink:1;flex-wrap:nowrap;float:none;font-family:initial;font-feature-settings:normal;font-kerning:auto;font-language-override:normal;font-size:medium;font-size-adjust:none;font-stretch:normal;font-style:normal;font-synthesis:weight style;font-variant:normal;font-variant-alternates:normal;font-variant-caps:normal;font-variant-east-asian:normal;font-variant-ligatures:normal;font-variant-numeric:normal;font-variant-position:normal;font-weight:400;grid-auto-columns:auto;grid-auto-flow:row;grid-auto-rows:auto;grid-column-end:auto;grid-column-gap:0;grid-column-start:auto;grid-row-end:auto;grid-row-gap:0;grid-row-start:auto;grid-template-areas:none;grid-template-columns:none;grid-template-rows:none;height:auto;hyphens:manual;image-orientation:0deg;image-rendering:auto;image-resolution:1dppx;ime-mode:auto;inline-size:auto;isolation:auto;justify-content:flexStart;left:auto;letter-spacing:normal;line-break:auto;line-height:normal;list-style-image:none;list-style-position:outside;list-style-type:disc;margin-block-end:0;margin-block-start:0;margin-bottom:0;margin-inline-end:0;margin-inline-start:0;margin-left:0;margin-right:0;margin-top:0;mask-clip:borderBox;mask-composite:add;mask-image:none;mask-mode:matchSource;mask-origin:borderBox;mask-position:0 0;mask-repeat:repeat;mask-size:auto;mask-type:luminance;max-height:none;max-width:none;min-block-size:0;min-height:0;min-inline-size:0;min-width:0;mix-blend-mode:normal;object-fit:fill;object-position:50% 50%;offset-block-end:auto;offset-block-start:auto;offset-inline-end:auto;offset-inline-start:auto;opacity:1;order:0;orphans:2;outline-color:initial;outline-offset:0;outline-style:none;outline-width:medium;overflow:visible;overflow-wrap:normal;overflow-x:visible;overflow-y:visible;padding-block-end:0;padding-block-start:0;padding-bottom:0;padding-inline-end:0;padding-inline-start:0;padding-left:0;padding-right:0;padding-top:0;page-break-after:auto;page-break-before:auto;page-break-inside:auto;perspective:none;perspective-origin:50% 50%;pointer-events:auto;position:static;quotes:initial;resize:none;right:auto;ruby-align:spaceAround;ruby-merge:separate;ruby-position:over;scroll-behavior:auto;scroll-snap-coordinate:none;scroll-snap-destination:0 0;scroll-snap-points-x:none;scroll-snap-points-y:none;scroll-snap-type:none;shape-image-threshold:0;shape-margin:0;shape-outside:none;tab-size:8;table-layout:auto;text-align:initial;text-align-last:auto;text-combine-upright:none;text-decoration-color:currentcolor;text-decoration-line:none;text-decoration-style:solid;text-emphasis-color:currentcolor;text-emphasis-position:over right;text-emphasis-style:none;text-indent:0;text-justify:auto;text-orientation:mixed;text-overflow:clip;text-rendering:auto;text-shadow:none;text-transform:none;text-underline-position:auto;top:auto;touch-action:auto;transform:none;transform-box:borderBox;transform-origin:50% 50%0;transform-style:flat;transition-delay:0s;transition-duration:0s;transition-property:all;transition-timing-function:ease;vertical-align:baseline;visibility:visible;white-space:normal;widows:2;width:auto;will-change:auto;word-break:normal;word-spacing:normal;word-wrap:normal;writing-mode:horizontalTb;z-index:auto;-webkit-appearance:none;-moz-appearance:none;-ms-appearance:none;appearance:none;margin:0}.LiveAreaSection-193358632{width:100%}.LiveAreaSection-193358632 .login-option-buybox{display:block;width:100%;font-size:17px;line-height:30px;color:#222;padding-top:30px;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-access-options{display:block;font-weight:700;font-size:17px;line-height:30px;color:#222;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-login>li:not(:first-child)::before{transform:translateY(-50%);content:””;height:1rem;position:absolute;top:50%;left:0;border-left:2px solid #999}.LiveAreaSection-193358632 .additional-login>li:not(:first-child){padding-left:10px}.LiveAreaSection-193358632 .additional-login>li{display:inline-block;position:relative;vertical-align:middle;padding-right:10px}.BuyBoxSection-683559780{display:flex;flex-wrap:wrap;flex:1;flex-direction:row-reverse;margin:-30px -15px 0}.BuyBoxSection-683559780 .box-inner{width:100%;height:100%}.BuyBoxSection-683559780 .readcube-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:1;flex-basis:255px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:300px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox-nature-plus{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:100%;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .title-readcube,.BuyBoxSection-683559780 .title-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .title-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .asia-link{color:#069;cursor:pointer;text-decoration:none;font-size:1.05em;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:1.05em6}.BuyBoxSection-683559780 .access-readcube{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;opacity:.8px;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .price-buybox{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;padding-top:30px;text-align:center}.BuyBoxSection-683559780 .price-buybox-to{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center}.BuyBoxSection-683559780 .price-info-text{font-size:16px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-value{font-size:30px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-per-period{font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-from{font-size:14px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .issue-buybox{display:block;font-size:13px;text-align:center;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:19px}.BuyBoxSection-683559780 .no-price-buybox{display:block;font-size:13px;line-height:18px;text-align:center;padding-right:10%;padding-left:10%;padding-bottom:20px;padding-top:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .vat-buybox{display:block;margin-top:5px;margin-right:20%;margin-left:20%;font-size:11px;color:#222;padding-top:10px;padding-bottom:15px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:17px}.BuyBoxSection-683559780 .tax-buybox{display:block;width:100%;color:#222;padding:20px 16px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:NaNpx}.BuyBoxSection-683559780 .button-container{display:flex;padding-right:20px;padding-left:20px;justify-content:center}.BuyBoxSection-683559780 .button-container>*{flex:1px}.BuyBoxSection-683559780 .button-container>a:hover,.Button-505204839:hover,.Button-1078489254:hover,.Button-2496381730:hover{text-decoration:none}.BuyBoxSection-683559780 .readcube-button{background:#fff;margin-top:30px}.BuyBoxSection-683559780 .button-asia{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;margin-top:75px}.BuyBoxSection-683559780 .button-label-asia,.ButtonLabel-3869432492,.ButtonLabel-3296148077,.ButtonLabel-1651148777{display:block;color:#fff;font-size:17px;line-height:20px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center;text-decoration:none;cursor:pointer}.Button-505204839,.Button-1078489254,.Button-2496381730{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;max-width:320px;margin-top:10px}.Button-505204839 .readcube-label,.Button-1078489254 .readcube-label,.Button-2496381730 .readcube-label{color:#069}
/* style specs end */
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout




References
-
Browne, A. J. et al. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planet. Health 5, e893–e904 (2021).
Google Scholar
-
Stewardson, A. J., Huttner, B. & Harbarth, S. At least it won’t hurt: the personal risks of antibiotic exposure. Curr. Opin. Pharmacol. 11, 446–452 (2011).
Google Scholar
-
Blaser, M. J. Antibiotic use and its consequences for the normal microbiome. Science 352, 544–545 (2016).
Google Scholar
-
Hogenauer, C., Hammer, H. F., Krejs, G. J. & Reisinger, E. C. Mechanisms and management of antibiotic‐associated diarrhea. Clin. Infect. Dis. 27, 702–710 (1998).
Google Scholar
-
McFarland, L. V. Epidemiology, risk factors and treatments for antibiotic-associated diarrhea. Dig. Dis. Basel Switz. 16, 292–307 (1998).
Google Scholar
-
Caballero-Flores, G., Pickard, J. M. & Núñez, G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-022-00833-7 (2022). A comprehensive review on the mechanisms and regulation of colonization resistance.
Google Scholar
-
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Google Scholar
-
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Google Scholar
-
Wypych, T. P. & Marsland, B. J. Antibiotics as instigators of microbial dysbiosis: implications for asthma and allergy. Trends Immunol. 39, 697–711 (2018).
Google Scholar
-
Yuan, J. et al. Long-term use of antibiotics and risk of type 2 diabetes in women: a prospective cohort study. Int. J. Epidemiol. 49, 1572–1581 (2020).
Google Scholar
-
Park, S. J. et al. Association between antibiotics use and diabetes incidence in a nationally representative retrospective cohort among Koreans. Sci. Rep. 11, 21681 (2021).
Google Scholar
-
Zhou, W. et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569, 663–671 (2019).
Google Scholar
-
Lynch, S. V. & Pedersen, O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016).
Google Scholar
-
Trasande, L. et al. Infant antibiotic exposures and early-life body mass. Int. J. Obes. 37, 16–23 (2013).
Google Scholar
-
Teng, C., Reveles, K. R., Obodozie-Ofoegbu, O. O. & Frei, C. R. Clostridium difficile infection risk with important antibiotic classes: an analysis of the FDA adverse event reporting system. Int. J. Med. Sci. 16, 630–635 (2019).
Google Scholar
-
Högenauer, C. et al. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N. Engl. J. Med. 355, 2418–2426 (2006).
Google Scholar
-
Shukla, A. & Sobel, J. D. Vulvovaginitis caused by Candida species following antibiotic exposure. Curr. Infect. Dis. Rep. 21, 44 (2019).
Google Scholar
-
Ben-Ami, R. et al. Antibiotic exposure as a risk factor for fluconazole-resistant Candida bloodstream infection. Antimicrob. Agents Chemother. 56, 2518–2523 (2012).
Google Scholar
-
Ichinohe, T. et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA 108, 5354–5359 (2011).
Google Scholar
-
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O. & Piddock, L. J. V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51 (2014).
Google Scholar
-
Bush, K. et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896 (2011).
Google Scholar
-
Yassour, M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81 (2016). This longitudinal study of the infant gut microbiome observed transient blooms of specific species and resistance levels during antibiotic treatment.
Google Scholar
-
Stecher, B., Maier, L. & Hardt, W.-D. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11, 277–284 (2013).
Google Scholar
-
Buffie, C. G. et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80, 62–73 (2012).
Google Scholar
-
Shen, Z. et al. Emerging carriage of NDM-5 and MCR-1 in Escherichia coli from healthy people in multiple regions in China: a cross sectional observational study. eClinicalMedicine 6, 11–20 (2018).
Google Scholar
-
van Hattem, J. M. et al. Prolonged carriage and potential onward transmission of carbapenemase-producing Enterobacteriaceae in Dutch travelers. Future Microbiol. 11, 857–864 (2016).
Google Scholar
-
Hu, Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4, 2151 (2013).
Google Scholar
-
Group, B. M. J. P. Superinfections during antibiotic treatment. Br. Med. J. 1, 537–538 (1952).
-
Ramirez, J. et al. Antibiotics as major disruptors of gut microbiota. Front. Cell. Infect. Microbiol. 10, 572912 (2020).
Google Scholar
-
Sullivan, Å. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1, 101–114 (2001). A review of the drug-specific effects of antibiotic on the microbiota, including extra-intestinal microbiota sites.
Google Scholar
-
Yang, L. et al. The varying effects of antibiotics on gut microbiota. AMB Express 11, 116 (2021).
Google Scholar
-
Maier, L. et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599, 120–124 (2021). This study comprehensively screened gut commensals to identify drug combinations active against pathogens but that minimize collateral damage against other species.
Google Scholar
-
Kelly, S. A., Rodgers, A. M., O’Brien, S. C., Donnelly, R. F. & Gilmore, B. F. Gut check time: antibiotic delivery strategies to reduce antimicrobial resistance. Trends Biotechnol. 38, 447–462 (2020).
Google Scholar
-
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA 108, 4554–4561 (2011). This study highlights the differences between individuals in the microbiota response and recovery to antibiotics.
Google Scholar
-
Jeffery, I. B., Lynch, D. B. & O’Toole, P. W. Composition and temporal stability of the gut microbiota in older persons. ISME J. 10, 170–182 (2016).
Google Scholar
-
Zimmermann, M., Patil, K. R., Typas, A. & Maier, L. Towards a mechanistic understanding of reciprocal drug–microbiome interactions. Mol. Syst. Biol. 17, e10116 (2021).
Google Scholar
-
Gjonbalaj, M. et al. Antibiotic degradation by commensal microbes shields pathogens. Infect. Immun. 88, e00012–e00020 (2020).
Google Scholar
-
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Google Scholar
-
Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 (2018).
Google Scholar
-
Elvers, K. T. et al. Antibiotic-induced changes in the human gut microbiota for the most commonly prescribed antibiotics in primary care in the UK: a systematic review. BMJ Open 10, e035677 (2020).
Google Scholar
-
Zimmermann, P. & Curtis, N. The effect of antibiotics on the composition of the intestinal microbiota — a systematic review. J. Infect. 79, 471–489 (2019).
Google Scholar
-
Levison, M. E. & Levison, J. H. Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect. Dis. Clin. North. Am. 23, 791–815 (2009).
Google Scholar
-
Levy, R. M., Huang, E. Y., Roling, D., Leyden, J. J. & Margolis, D. J. Effect of antibiotics on the oropharyngeal flora in patients with acne. Arch. Dermatol. 139, 467–471 (2003).
Google Scholar
-
Kim, S., Covington, A. & Pamer, E. G. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 279, 90–105 (2017).
Google Scholar
-
Kelly, S. A. et al. Antibiotic therapy and the gut microbiome: investigating the effect of delivery route on gut pathogens. ACS Infect. Dis. 7, 1283–1296 (2021).
Google Scholar
-
Zhang, L., Huang, Y., Zhou, Y., Buckley, T. & Wang, H. H. Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother. 57, 3659–3666 (2013). A comparison of oral and intravenous antibiotic administration on the spread of antibiotic resistance in the mouse intestine.
Google Scholar
-
Finegold, S. M. Anaerobic infections in humans: an overview. Anaerobe 1, 3–9 (1995).
Google Scholar
-
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Google Scholar
-
Donskey, C. J. et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N. Engl. J. Med. 343, 1925–1932 (2000). This study of patients colonized with vancomycin-resistant enterococci showed overgrowth in the intestine during treatment with various anti-anaerobic antibiotics.
Google Scholar
-
Brook, I., Wexler, H. M. & Goldstein, E. J. C. Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin. Microbiol. Rev. 26, 526–546 (2013).
Google Scholar
-
Taur, Y. et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914 (2012). Intestinal domination by various bacteria is associated with subsequent bacteraemia in patients undergoing haematopoietic stem cell transplantation.
Google Scholar
-
Young, V. B. & Schmidt, T. M. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J. Clin. Microbiol. 42, 1203–1206 (2004).
Google Scholar
-
Wiström, J. et al. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: a prospective study. J. Antimicrob. Chemother. 47, 43–50 (2001).
Google Scholar
-
Ma, H. et al. Combined administration of antibiotics increases the incidence of antibiotic-associated diarrhea in critically ill patients. Infect. Drug. Resist. 12, 1047–1054 (2019).
Google Scholar
-
Rashidi, A. et al. Gut microbiota response to antibiotics is personalized and depends on baseline microbiota. Microbiome 9, 211 (2021).
Google Scholar
-
Arvidsson, A., Leijd, B., Nord, C. E. & Angelin, B. Interindividual variability in biliary excretion of ceftriaxone: effects on biliary lipid metabolism and on intestinal microflora. Eur. J. Clin. Invest. 18, 261–266 (1988).
Google Scholar
-
Coyte, K. Z., Schluter, J. & Foster, K. R. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666 (2015).
Google Scholar
-
Dethlefsen, L., Huse, S., Sogin, M. L. & Relman, D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280 (2008).
Google Scholar
-
Anthony, W. E. et al. Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep. 39, 110649 (2022). This study shows short- and long-term effects of antibiotics on the gut microbiota of healthy volunteers.
Google Scholar
-
Yee, A. L. et al. Longitudinal microbiome composition and stability correlate with increased weight and length of very-low-birth-weight infants. mSystems 4, e00229-18 (2019).
Google Scholar
-
Sorg, R. A. et al. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLoS Biol. 14, e2000631 (2016).
Google Scholar
-
Cubillos-Ruiz, A. et al. An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nat. Biomed. Eng. 6, 910–921 (2022).
Google Scholar
-
Sun, J. et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 11, 1427 (2020).
Google Scholar
-
Foster, K. R., Schluter, J., Coyte, K. Z. & Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017).
Google Scholar
-
Schirmer, M. et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167, 1125–1136.e8 (2016).
Google Scholar
-
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Google Scholar
-
Montandon, S. A. & Jornayvaz, F. R. Effects of antidiabetic drugs on gut microbiota composition. Genes 8, 250 (2017).
Google Scholar
-
Le Bastard, Q. et al. Systematic review: human gut dysbiosis induced by non-antibiotic prescription medications. Aliment. Pharmacol. Ther. 47, 332–345 (2018).
Google Scholar
-
Kwok, C. S. et al. Risk of Clostridium difficile infection with acid suppressing drugs and antibiotics: meta-analysis. Am. J. Gastroenterol. 107, 1011–1019 (2012).
Google Scholar
-
Trifan, A. et al. Proton pump inhibitors therapy and risk of Clostridium difficile infection: systematic review and meta-analysis. World J. Gastroenterol. 23, 6500–6515 (2017).
Google Scholar
-
Caballero-Flores, G., Pickard, J. M., Fukuda, S., Inohara, N. & Núñez, G. An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe 28, 526–533.e5 (2020).
Google Scholar
-
Theriot, C. M. et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5, 3114 (2014).
Google Scholar
-
Sassone-Corsi, M. et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016).
Google Scholar
-
Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
Google Scholar
-
Ivanov, I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Google Scholar
-
Deshmukh, H. S. et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20, 524–530 (2014).
Google Scholar
-
Pamer, E. G. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352, 535–538 (2016).
Google Scholar
-
Freter, R. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J. Infect. Dis. 97, 57–65 (1955). An early study identifying the protection against infection conferred by an intact microbiota.
Google Scholar
-
Miller, C. P., Bohnhoff, M. & Rifkind, D. The effect of an antibiotic on the susceptibility of the mouse’s intestinal tract to Salmonella infection. Trans. Am. Clin. Climatol. Assoc. 68, 51–58 (1957).
Google Scholar
-
Sekirov, I. et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 (2008).
Google Scholar
-
Hensgens, M. P. M., Goorhuis, A., Dekkers, O. M. & Kuijper, E. J. Time interval of increased risk for Clostridium difficile infection after exposure to antibiotics. J. Antimicrob. Chemother. 67, 742–748 (2012). A multicenter case–control study to determine the period at risk for CDI after cessation of antibiotics.
Google Scholar
-
Tvede, M. & Rask-Madsen, J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 333, 1156–1160 (1989).
-
van Nood, E. et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415 (2013).
Google Scholar
-
Buffie, C. G. et al. Precision microbiome restoration of bile acid-mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). Bile-acid-mediated colonization resistance against C. difficile could be restored by the human gut commensal C. scindens.
Google Scholar
-
Aguirre, A. M. et al. Bile acid-independent protection against Clostridioides difficile infection. PLoS Pathog. 17, e1010015 (2021).
Google Scholar
-
Gregory, A. L., Pensinger, D. A. & Hryckowian, A. J. A short chain fatty acid-centric view of Clostridioides difficile pathogenesis. PLoS Pathog. 17, e1009959 (2021).
Google Scholar
-
Iwata, K. et al. A systematic review for pursuing the presence of antibiotic associated enterocolitis caused by methicillin resistant Staphylococcus aureus. BMC Infect. Dis. 14, 247 (2014).
Google Scholar
-
Lane, A. B., Copeland, N. K., Onmus-Leone, F. & Lawler, J. V. Methicillin-resistant Staphylococcus aureus as a probable cause of antibiotic-associated enterocolitis. Case Rep. Infect. Dis. 2018, e3106305 (2018).
-
Lichtman, J. S. et al. Host–microbiota interactions in the pathogenesis of antibiotic-associated diseases. Cell Rep. 14, 1049–1061 (2016).
Google Scholar
-
Pavia, A. T. et al. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J. Infect. Dis. 161, 255–260 (1990).
Google Scholar
-
Holmberg, S. D., Osterholm, M. T., Senger, K. A. & Cohen, M. L. Drug-resistant Salmonella from animals fed antimicrobials. N. Engl. J. Med. 311, 617–622 (1984).
Google Scholar
-
Gradel, K. O., Dethlefsen, C., Ejlertsen, T., Schønheyder, H. C. & Nielsen, H. Increased prescription rate of antibiotics prior to non-typhoid Salmonella infections: a one-year nested case–control study. Scand. J. Infect. Dis. 40, 635–641 (2008).
Google Scholar
-
Doorduyn, Y., Van Den Brandhof, W. E., Van Duynhoven, Y. T. H. P., Wannet, W. J. B. & Van Pelt, W. Risk factors for Salmonella Enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiol. Infect. 134, 617–626 (2006).
Google Scholar
-
Malik, U. et al. Association between prior antibiotic therapy and subsequent risk of community-acquired infections: a systematic review. J. Antimicrob. Chemother. 73, 287–296 (2018).
Google Scholar
-
Humphreys, H. et al. Four country healthcare associated infection prevalence survey 2006: risk factor analysis. J. Hosp. Infect. 69, 249–257 (2008).
Google Scholar
-
Chen, Y. E., Fischbach, M. A. & Belkaid, Y. Skin microbiota–host interactions. Nature 553, 427–436 (2018).
Google Scholar
-
Nakatsuji, T. et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680 (2017).
Google Scholar
-
Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016).
Google Scholar
-
Belkaid, Y. & Segre, J. A. Dialogue between skin microbiota and immunity. Science 346, 954–959 (2014).
Google Scholar
-
Liu, Q. et al. Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 27, 68–78.e5 (2020).
Google Scholar
-
Man, W. H., de Steenhuijsen Piters, W. A. A. & Bogaert, D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270 (2017).
Google Scholar
-
Clark, S. E. Commensal bacteria in the upper respiratory tract regulate susceptibility to infection. Curr. Opin. Immunol. 66, 42–49 (2020).
Google Scholar
-
Santagati, M., Scillato, M., Patanè, F., Aiello, C. & Stefani, S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol. Med. Microbiol. 65, 23–31 (2012).
Google Scholar
-
Horn, K. J. et al. Corynebacterium species inhibit Streptococcus pneumoniae colonization and infection of the mouse airway. Front. Microbiol. 12, 804935 (2021).
Google Scholar
-
Thackray, L. B. et al. Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 22, 3440–3453.e6 (2018).
Google Scholar
-
Margolis, D. J., Bowe, W. P., Hoffstad, O. & Berlin, J. A. Antibiotic treatment of acne may be associated with upper respiratory tract infections. Arch. Dermatol. 141, 1132–1136 (2005).
Google Scholar
-
Smith, H. S. et al. Antecedent antimicrobial use increases the risk of uncomplicated cystitis in young women. Clin. Infect. Dis. 25, 63–68 (1997).
Google Scholar
-
Robinson, C. J. & Young, V. B. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes 1, 279–284 (2010).
Google Scholar
-
Zacharioudakis, I. M., Zervou, F. N., Pliakos, E. E., Ziakas, P. D. & Mylonakis, E. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: a systematic review and meta-analysis. Am. J. Gastroenterol. 110, 381–390 (2015).
Google Scholar
-
Stevens, E. J., Bates, K. A. & King, K. C. Host microbiota can facilitate pathogen infection. PLoS Pathog. 17, e1009514 (2021).
Google Scholar
-
Kluytmans-van den Bergh, M. F. Q. et al. Rectal carriage of extended-spectrum-β-lactamase-producing enterobacteriaceae in hospitalized patients: selective preenrichment increases yield of screening. J. Clin. Microbiol. 53, 2709–2712 (2015).
Google Scholar
-
Sheppard, S. K. Strain wars and the evolution of opportunistic pathogens. Curr. Opin. Microbiol. 67, 102138 (2022).
Google Scholar
-
Donskey, C. J. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39, 219–226 (2004).
Google Scholar
-
Sim, C. K. et al. A mouse model of occult intestinal colonization demonstrating antibiotic-induced outgrowth of carbapenem-resistant Enterobacteriaceae. Microbiome 10, 43 (2022). Mice colonized with resistant bacteria at undetectable levels showed overgrowth following antibiotic treatment.
Google Scholar
-
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013).
Google Scholar
-
Bottery, M. J. et al. Inter-species interactions alter antibiotic efficacy in bacterial communities. ISME J. 16, 812–821 (2022).
Google Scholar
-
Tavernier, S. et al. Community composition determines activity of antibiotics against multispecies biofilms. Antimicrob. Agents Chemother. 61, e00302–e00317 (2017).
Google Scholar
-
Brown, G. D. et al. Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv13 (2012).
Google Scholar
-
Mayer, F. L., Wilson, D. & Hube, B. Candida albicans pathogenicity mechanisms. Virulence 4, 119–128 (2013).
Google Scholar
-
Xu, J. et al. Effect of antibiotics on vulvovaginal candidiasis: a MetroNet study. J. Am. Board. Fam. Med. 21, 261–268 (2008).
Google Scholar
-
MacDonald, T. M. et al. The risks of symptomatic vaginal candidiasis after oral antibiotic therapy. Q. J. Med. 86, 419–424 (1993).
Google Scholar
-
Tan, C. T., Xu, X., Qiao, Y. & Wang, Y. A peptidoglycan storm caused by β-lactam antibiotic’s action on host microbiota drives Candida albicans infection. Nat. Commun. 12, 2560 (2021).
Google Scholar
-
Seelig, M. S. The role of antibiotics in the pathogenesis of Candida infections. Am. J. Med. 40, 887–917 (1966).
Google Scholar
-
Takahashi, S. et al. Septic pulmonary embolism caused by Candida albicans after treatment for urinary multidrug-resistant Pseudomonas aeruginosa. J. Infect. Chemother. 14, 436–438 (2008).
Google Scholar
-
Zhai, B. et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat. Med. 26, 59–64 (2020).
Google Scholar
-
Samonis, G. et al. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob. Agents Chemother. 37, 51–53 (1993).
Google Scholar
-
Spigaglia, P., Mastrantonio, P. & Barbanti, F. in Updates on Clostridium difficile in Europe: Advances in Microbiology, Infectious Diseases and Public Health Volume 8 (eds Mastrantonio, P. & Rupnik, M.) 137–159 (Springer International, 2018). https://doi.org/10.1007/978-3-319-72799-8_9.
-
Toth, M., Stewart, N. K., Smith, C. & Vakulenko, S. B. Intrinsic class D β-lactamases of Clostridium difficile. mBio 9, e01803–e01818 (2018).
Google Scholar
-
Eyre, D. W. et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 369, 1195–1205 (2013). A study showing that many C. difficile infections are not the result of transmission chains in hospital settings.
Google Scholar
-
Ayres, J. S., Trinidad, N. J. & Vance, R. E. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 18, 799–806 (2012).
Google Scholar
-
Segura Munoz, R. R. et al. Experimental evaluation of ecological principles to understand and modulate the outcome of bacterial strain competition in gut microbiomes. ISME J. 16, 1594–1604 (2022).
Google Scholar
-
Hardin, G. The competitive exclusion principle. Science 131, 1292–1297 (1960).
Google Scholar
-
Lentsch, V. et al. Combined oral vaccination with niche competition can generate sterilizing immunity against enteropathogenic bacteria. Preprint at bioRxiv https://doi.org/10.1101/2022.07.20.498444 (2022).
-
Lee, S. M. et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013).
Google Scholar
-
Murray, B. E., Rensimer, E. R. & Dupont, H. L. Emergence of high-level trimethoprim resistance in fecal Escherichia coli during oral administration of trimethoprim or trimethoprim–sulfamethoxazole. N. Engl. J. Med. 306, 130–135 (1982).
Google Scholar
-
Palleja, A. et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 3, 1255–1265 (2018).
Google Scholar
-
Vollaard, E. J., Clasener, H. A. L., van Griethuysen, A. J. A., Janssen, A. J. & Sanders-Reijmers, A. J. Influence of amoxycillin, erythromycin and roxithromycin on colonization resistance and on appearance of secondary colonization in healthy volunteers. J. Antimicrob. Chemother. 20, 131–138 (1987).
Google Scholar
-
Brandl, K. et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807 (2008).
Google Scholar
-
Ubeda, C. et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010). This study showed how VRE overgrow in the intestine during antibiotic treatment.
Google Scholar
-
Soares, F. S. et al. Antibiotic-induced pathobiont dissemination accelerates mortality in severe experimental pancreatitis. Front. Immunol. 8, 1890 (2017).
Google Scholar
-
Drummond, R. A. et al. Long-term antibiotic exposure promotes mortality after systemic fungal infection by driving lymphocyte dysfunction and systemic escape of commensal bacteria. Cell Host Microbe 30, 1020–1033.e6 (2022).
Google Scholar
-
Knoop, K. A., McDonald, K. G., Kulkarni, D. H. & Newberry, R. D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 65, 1100–1109 (2016).
Google Scholar
-
Forde, B. M. et al. Population dynamics of an Escherichia coli ST131 lineage during recurrent urinary tract infection. Nat. Commun. 10, 3643 (2019).
Google Scholar
-
Flores-Mireles, A. L., Walker, J. N., Caparon, M. & Hultgren, S. J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284 (2015).
Google Scholar
-
Wheatley, R. M. et al. Gut to lung translocation and antibiotic mediated selection shape the dynamics of Pseudomonas aeruginosa in an ICU patient. Nat. Commun. 13, 6523 (2022).
Google Scholar
-
Lawley, T. D. et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77, 3661–3669 (2009).
Google Scholar
-
Magill, S. S. et al. Prevalence of antimicrobial use in US acute care hospitals, May–September 2011. J. Am. Med. Asssoc. 312, 1438–1446 (2014).
Google Scholar
-
Stracy, M. et al. Minimizing treatment-induced emergence of antibiotic resistance in bacterial infections. Science 375, 889–894 (2022). Personalized antibiotic recommendations could reduce the emergence of resistance during antibiotic treatment.
Google Scholar
-
Caballero, J. D. et al. Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients. Nat. Commun. 14, 4083 (2023).
-
Tchesnokova, V. L. et al. Pandemic uropathogenic fluoroquinolone-resistant Escherichia coli have enhanced ability to persist in the gut and cause bacteriuria in healthy women. Clin. Infect. Dis. 70, 937–939 (2020).
Google Scholar
-
von Eiff, C., Becker, K., Machka, K., Stammer, H. & Peters, G. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344, 11–16 (2001).
-
Gasparrini, A. J. et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 4, 2285–2297 (2019).
Google Scholar
-
Korpela, K. et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7, 10410 (2016).
Google Scholar
-
Jernberg, C., Löfmark, S., Edlund, C. & Jansson, J. K. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol. Read. Engl. 156, 3216–3223 (2010).
Google Scholar
-
Wenzler, E., Mulugeta, S. G. & Danziger, L. H. The antimicrobial stewardship approach to combating Clostridium difficile. Antibiotics 4, 198–215 (2015).
Google Scholar
-
Baur, D. et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect. Dis. 17, 990–1001 (2017).
Google Scholar
-
Aldeyab, M. A. et al. An evaluation of the impact of antibiotic stewardship on reducing the use of high-risk antibiotics and its effect on the incidence of Clostridium difficile infection in hospital settings. J. Antimicrob. Chemother. 67, 2988–2996 (2012).
Google Scholar
-
Wiesch, P. A., zur, Kouyos, R., Abel, S., Viechtbauer, W. & Bonhoeffer, S. Cycling empirical antibiotic therapy in hospitals: meta-analysis and models. PLoS Pathog. 10, e1004225 (2014).
-
Moser, C. et al. Antibiotic therapy as personalized medicine — general considerations and complicating factors. APMIS 127, 361–371 (2019).
Google Scholar
-
Yeh, Y.-C., Huang, T.-H., Yang, S.-C., Chen, C.-C. & Fang, J.-Y. Nano-based drug delivery or targeting to eradicate bacteria for infection mitigation: a review of recent advances. Front. Chem. 8, 286 (2020).
Google Scholar
-
Wang, Y. et al. Targeted delivery of antibiotics to the infected pulmonary tissues using ROS-responsive nanoparticles. J. Nanobiotechnol. 17, 103 (2019).
-
Yao, J. et al. A pathogen-selective antibiotic minimizes disturbance to the microbiome. Antimicrob. Agents Chemother. 60, 4264–4273 (2016).
Google Scholar
-
Mu, H. et al. Pathogen-targeting glycovesicles as a therapy for salmonellosis. Nat. Commun. 10, 4039 (2019).
Google Scholar
-
Brochado, A. R. et al. Species-specific activity of antibacterial drug combinations. Nature 559, 259–263 (2018).
Google Scholar
-
Gutiérrez, B. & Domingo-Calap, P. Phage therapy in gastrointestinal diseases. Microorganisms 8, 1420 (2020).
Google Scholar
-
Cotter, P. D., Ross, R. P. & Hill, C. Bacteriocins — a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105 (2013).
Google Scholar
-
Meade, E., Slattery, M. A. & Garvey, M. Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: resistance is futile? Antibiotics 9, 32 (2020).
Google Scholar
-
Hatfull, G. F., Dedrick, R. M. & Schooley, R. T. Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211 (2022).
Google Scholar
-
Dedrick, R. M. et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019).
Google Scholar
-
Schooley, R. T. et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954-17 (2017).
Google Scholar
-
Rea, M. C. et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl Acad. Sci. USA 107, 9352–9357 (2010).
Google Scholar
-
McDonald, L. C. et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 66, 987–994 (2018).
Google Scholar
-
Yelin, I. et al. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat. Med. 25, 1728–1732 (2019).
Google Scholar
-
Suez, J. et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.e16 (2018).
Google Scholar
-
Imperial, I. C. V. J. & Ibana, J. A. Addressing the antibiotic resistance problem with probiotics: reducing the risk of its double-edged sword effect. Front. Microbiol. 7, 1983 (2016).
Google Scholar
-
Kassam, Z., Lee, C. H., Yuan, Y. & Hunt, R. H. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol. 108, 500 (2013).
Google Scholar
-
Pamer, E. G. Fecal microbiota transplantation: effectiveness, complexities, and lingering concerns. Mucosal Immunol. 7, 210–214 (2014).
Google Scholar
-
Taur, Y. et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med. 10, eaap9489 (2018).
Google Scholar
-
DeFilipp, Z. et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Google Scholar
-
Amrane, S. & Lagier, J.-C. Fecal microbiota transplantation for antibiotic resistant bacteria decolonization. Hum. Microbiome J. 16, 100071 (2020). A comprehensive overview of the application of FMT to decolonize the gut of antibiotic resistance bacteria.
-
Walker, A. W. et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5, 220–230 (2011).
Google Scholar
-
Stecher, B. & Hardt, W.-D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 14, 82–91 (2011).
Google Scholar
-
Pickard, J. M. & Núñez, G. Pathogen colonization resistance in the gut and its manipulation for improved health. Am. J. Pathol. 189, 1300–1310 (2019).
Google Scholar
-
Russell, A. B., Peterson, S. B. & Mougous, J. D. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014).
Google Scholar
-
Brunet, Y. R., Espinosa, L., Harchouni, S., Mignot, T. & Cascales, E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 3, 36–41 (2013).
Google Scholar
-
Cotter, P. D., Hill, C. & Ross, R. P. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788 (2005).
Google Scholar
-
Vital, M., Rud, T., Rath, S., Pieper, D. H. & Schlüter, D. Diversity of bacteria exhibiting bile acid-inducible 7α-dehydroxylation genes in the human gut. Comput. Struct. Biotechnol. J. 17, 1016–1019 (2019).
Google Scholar
-
Didelot, X., Walker, A. S., Peto, T. E., Crook, D. W. & Wilson, D. J. Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 14, 150–162 (2016).
Google Scholar
-
Ikuta, K. S. et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 400, 2221–2248 (2022).
-
Magruder, M. et al. Gut uropathogen abundance is a risk factor for development of bacteriuria and urinary tract infection. Nat. Commun. 10, 5521 (2019).
Google Scholar
Acknowledgements
This research was funded by Wellcome Trust grant 224212/Z/21/Z.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article. L.d.N. and M.S. contributed substantially to discussion of the content. L.d.N. and M.S. wrote the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Simone Becattini and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and Permissions
About this article
Cite this article
de Nies, L., Kobras, C.M. & Stracy, M. Antibiotic-induced collateral damage to the microbiota and associated infections.
Nat Rev Microbiol (2023). https://doi.org/10.1038/s41579-023-00936-9
-
Accepted: 28 June 2023
-
Published: 04 August 2023
-
DOI: https://doi.org/10.1038/s41579-023-00936-9