Cancer and neoplasms
Analysis of endoplasmic reticulum stress-related gene signature for the prognosis and pattern in diffuse large B cell lymphoma
Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoma in adults. This study aimed to determine the prognostic significance of endoplasmic reticulum (ER) stress-related genes in DLBCL. ER stress-related genes were obtained from the molecular signatures database. Gene expression data and clinical outcomes from the gene expression omnibus and TCGA datasets were collected, and differentially expressed genes (DEGs) were screened out. Gene ontology enrichment analysis, the kyoto encyclopaedia of genes and genomes pathway analysis, and geneset enrichment analysis were used to analyse the possible biological function of ER stress-related DEGs in DLBCL. Protein–protein interaction network construction using the STRING online and hub genes were identified by cytoHubba on Cytoscape software. The significant prognosis-related genes were screened, and the differential expression was validated. The immune microenvironment assessment of significant genes were evaluated. Next, the nomogram was built using univariate and multivariate Cox regression analysis. 26 ER stress-related DEGs were screened. Functional enrichment analysis showed them to be involved in the regulation of the endoplasmic reticulum mainly. NUPR1 and TRIB3 were identified as the most significant prognostic-related genes by comparison with the GSE10846, GSE11318, and TCGA datasets. NUPR1 was correlated with a good prognosis and immune infiltration in DLBCL; on the other hand, high expression of TRIB3 significantly correlated with a poor prognosis, which was an independent prognostic factor for DLBCL. In summary, we identified NUPR1 and TRIB3 as critical ER stress-related genes in DLBCL. NUPR1 might be involved in immune infiltration in DLBCL, and TRIB3 might serve as a potential therapeutic target and prognostic factor in DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid neoplasm, representing approximately 30% of non-Hodgkin lymphoma (NHL)1,2, and it is a highly heterogeneous, aggressive disease2,3. Currently, R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) is the standard first-line treatment for DLBCL2,4, however, about 30–40% of DLBCL patients remain resistant to RCHOP and are refractory or develop relapsed5,6, and only 10% of patients with refractory or relapsed DLBCL could be treated using salvage immunochemotherapy followed by autologous stem cell transplantation3,7, implying a significant unmet medical need8,9. The cell-of-origin (COO) classifications9,10 and the International Prognostic Index (IPI) score have often been considered the two most commonly prognostic factors for patients with DLBCL3,11,12. However, these prognostic factors do not fully explain risk stratification, clinical outcomes in DLBCL patients13,14, and there is an urgent need for valuable biomarkers to guide prognostic factors and therapeutic approaches for DLBCL.
Endoplasmic reticulum (ER) stress, a state in which the unfolded and misfolded protein accumulation affects the normal physiological function of cells, refers to the excessive stress caused by dysfunction of ER stress15,16. Notably, ER stress signaling is associated with the development of several cancers, including DLBCL15,16,17. The unfolded protein response (UPR), controlled general translation, misfolded protein degradation, and folding enzyme production are some of the adaptive responses that cells may initiate in response to ER stress. Protein kinase RNA (PKR)-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1) are the three types of signal transducers evolved to be involved in the regulation of UPR network18,19. Malignant and stromal cells have their ER homeostasis disrupted by the hostile milieu created by a confluence of oncogenic, transcriptional, and metabolic aberrations in numerous tumor forms18,20,21. The alterations induce a state of persistent ER stress, which has been shown to regulate several protumor characteristics in cancer cells while dynamically altering the function of innate and adaptive immune cells. Generally, aberrant activation of ER stress sensors and their downstream signalling pathways have been identified as important regulators of cancer development, metastasis, and response to chemotherapy, targeted treatments, and immunotherapy19,20,21,22,23. But the significance of ER stress-related genes in the biological features and clinical prognosis of DLBCL has not been thoroughly investigated.
In our study, we aimed to better investigate and comprehend the predictive and prognostic value of ER stress-related genes in DLBCL using public databases. First, differentially expressed genes (DEGs) were compared in DLBCL patients with normal tissues. Second, the biological activities and potential pathways of ER stress-related DEGs were validated. Third, the prognostic model and immune infiltration assessment were performed. We explored and verified to illustrate the prognostic role and biological functions of ER stress-related genes in DLBCL at the bioinformatic and experimental levels.
Materials and methods
Data downloading and preprocessing
DLBCL datasets (GSE5631524, GSE1084625, and GSE1131826) were obtained from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). All three datasets were based on the GPL570 platform and came from Homo sapiens. There were 55 DLBCL patients and 33 noncancerous tissues (NCs) in the GSE56315 dataset, and all of them were included in this analysis. After removing patients with incomplete survival information, 414 DLBCL samples in the GSE10846 dataset and 200 DLBCL samples in the GSE11318 dataset were enrolled. And 48 DLBCL samples (TCGA_DLBC) in The Cancer Genome Atlas (TCGA) were downloaded from The University of California Santa Cruz (UCSC) Xena browser. A total of 295 ER stress-related geneset were obtained from the Molecular Signature Database v7.0 (MSigDB)27 after removing the overlapped genes (Supplementary Table S1).
DEGs screening and gene function analysis
DEGs had been screened between DLBCL samples and lymphocytal data from normal human tonsils, which came from the GSE56315 dataset, and were explored using the limma package28. Two thresholds were set to determine the degree of DEGs: the adjusted p value 1. And these DEGs were intersected with the ER stress-related genesets to extract the differentially ER stress-related DEGs. For the exploration of the function of the extracted genes, the gene ontology (GO) enrichment analysis29, and kyoto encyclopedia of genes and genomes (KEGG) pathway analysis30 were conducted using the clusterProfiler package31. In order to evaluate the gene expression level of potential signaling pathways and biological functions, the gene set enrichment analysis (GSEA)32 was conducted using the “c2.cp.kegg.v6.2.-symbols” geneset, and false discovery rate (FDR) q-value ≤ 0.25 was considered as statistically significant.
Protein–protein interaction network construction
For searching for the relationship between proteins of interest, a Protein–protein interaction (PPI) network was constructed through the Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org)33 online, and the combined score was greater than 0.4 considered statistically significant. Cytoscape (version 3.7.2, http://www.cytoscape.org)34 was used to visualize this PPI network, and the cytoHubba plug-in Cytoscape35 was used to calculate the hub nodes to select the top DEGs based on degree and the maximum correntropy criterion (MCC) algorithms.
Screening of prognosis-related genes and clinical correlation analysis
The GSE10846 dataset25, the GSE11318 dataset26, and the TCGA_DLBC databases were used to discover prognosis-related genes in DLBCL patients, and the clinical characteristics were extracted. For the investigation of the significant prognostic genes of ER stress-related DEGs, the samples of above the three datasets were divided into two groups based on the median value of differential ER stress-related DEGs expression and the intersection of them. The Kaplan–Meier (KM) curves were employed for survival analysis, and the log-rank method was used to compare the two groups, P < 0.05 was considered as significant difference. Simultaneously, the expression of ER stress-related DEGs in different COO and stages of DLBCL was analyzed by Kruskal Wallis test. In order to investigate the diagnostic role of ER stress-related DEGs in the GSE56315 dataset, the receiver operating characteristic (ROC) curve was drawn through the GSE56315 dataset using the pROC and plotROC packages36, and the area under the curve (AUC) was calculated. The experssion of ER stress-related DEGs in the TCGA_DLBC was also determined.
Gene expression quantification on cultured cell lines
In this study, we detected gene expression in cultured cell lines, there are four cell lines including HBL-1 (A gift from Fujian Research Institute of Haematology, China), SUDHL2 (A gift from School of Medcine, Southeast Universtiy, China), SUDHL4 (Meisen, China) and GM12878 (BeNa, China) enrolled, thess cell lines were cultured in Roswell Park Memorial Institute 1640 (RPMI‐1640) medium (Gibco, US), supplemented with 10% fetal bovine serum (FBS, Gibco, US) and 1% Penicillin/Streptomycin (Gibco, US), and incubated at 37℃ in a 5% CO2 incubator. Total RNA were isolated using TRIzol reagent (Invitrogen, US), then HiScript® Q RT SuperMix for qPCR (Vazyme, China) was used to transcribe the RNA into cDNA. The Quantitative real‐time polymerase chain reaction (qRT-PCR) was performed on the CFX Connect Real-Time PCR Detection System (BioRad, US) with an HQ SYBR qPCR Mix (Without ROX) (Zomen, China). The mRNA expressions were quantified with the 2−ΔΔCt method, and β-actin expression was used as an endogenous reference. The primer sequences of identified ER stress-related DEGs were purchased from Sunya, China (Supplementary Table S2).
Immune microenvironment assessment
For evaluating the amount and proportion of the expression of ER stress-related DEGs in immune cells, CIBERSORTx37,38, a suite of machine learning tools that used the deconvolution algorithm to evaluate the proportion of various immune cells from the expression profiles of tumor tissues, and leukocyte signature matrix (LM22) feature matrices were used to estimate the proportion of immune cell in the DLBCL samples in the GSE10846 and GSE11318 datasets using the ESTIMATE package39, which provides the algorithm to calculate the tumor purity, stromal score, immune score.
Construction and validation of the nomogram based on the prognostic model
A nomogram integrating the expression of ER stress-related DEGs and clinical characteristics of DLBCL patients, including tumor-node-metastasis (TNM) stage, Eastern Cooperative Oncology Group (ECOG) score, lactate dehydrogenase (LDH) ratio, and others, was constructed for prediction of prognosis in the GSE10846 as a training datasets. Furthermore, the nomogram’s prognosis prediction performance was confirmed in the GSE11318 datasets (validation dataset) by comparing the fitting degree between the observed and optimized values.
Statistical analysis
R programming (version 4.1) was used for all statistical analysis. We used the Student’s t-test or one-way analysis of variance (ANOVA) to compare normally distributed continuous variables between groups and Mann–Whitney test or Kruskal–Wallis test to examine non-normally distributed continuous variables. Two-sided P values less than 0.05 indicated statistical significance.
Results
Screening of ER stress-related DEGs
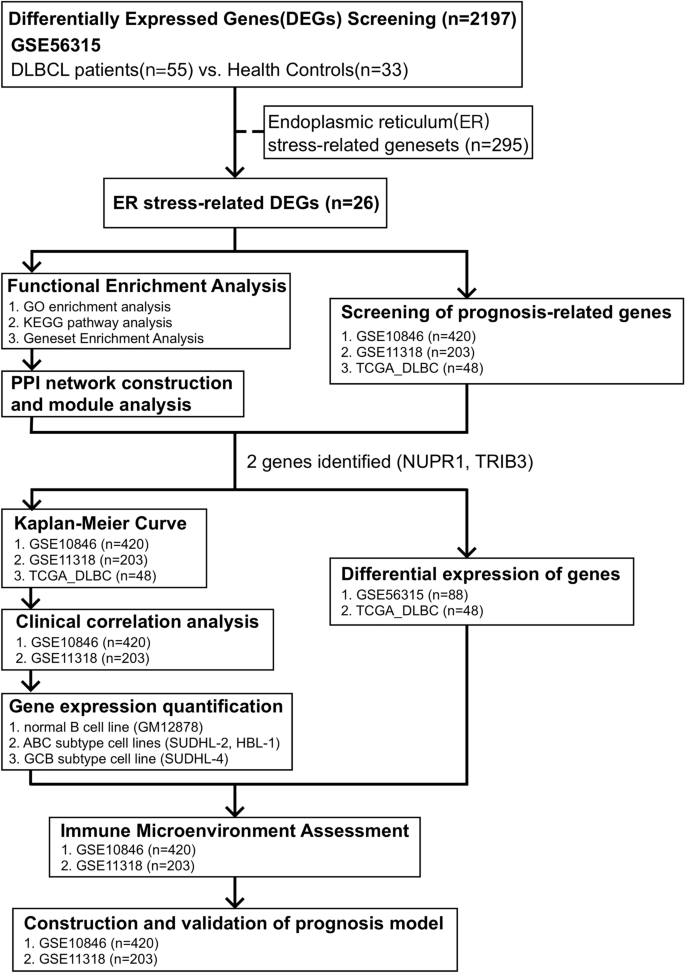
This study’s flowchart is depicted in Fig. 1. The clinical characteristics and standardized gene data were extracted from the GSE56315 dataset using GEOquery package40, and there are significant differences between DLBCL and NCs based on the principal component analysis (PCA), as shown in Fig. 2A. A total of 2179 DEGs were obtained through difference analysis using the limma package28, among which 1367 genes were upregulated and 812 genes were downregulated (Fig. 2B,C). After determining the overlap between ER stress-related genesets and DEGs through a Venn diagram, 26 ER stress-related DEGs were found (Fig. 2D).
Workflow of this study.

Identification of ER stress-related DEGs. (A) PCA show the dimensionality reduction distribution of DLBCL samples and NCs in the GSE56315 dataset. (B)–(C) Heatmap plot and volcano plot of DEGs in DLBCL samples vs. NCs based on the GSE56315 dataset. (D) Venn diagram of ER stress-related genes and DEGs. DLBCL Diffuse Large B Cell Lymphoma, DEGs differentially expressed genes, ER endoplasmic reticulum, PCA principal component analysis.
Functional enrichment analysis and PPI network construction
In order to find the enriched functions for the 26 ER stress-related DEGs, GO enrichment analysis were processed. The significantly enriched biological processes (BP) included response to ER stress, cellular response to stress, and response to stress. And the main top enrichment cellular component (CC) was ER (Fig. 3A). The KEGG pathway analysis showed that protein processing in ER, fluid shear stress, tumor necrosis factor (TNF) signaling pathway, atherosclerosis, and insulin resistance were enriched (Fig. 3B). More details of the top 50 significant items of GO enrichment analysis and KEGG pathway analysis (Ref: 231,102) could be shown in Supplementary Tables S3 and S4. The above results suggested that the metabolism and immune system were critical for DLBCL. We carried out a GSEA analysis to explore the metabolism and immune-related pathways; there were some pathways, including amino sugar and nucleotide sugar metabolism, diseases of metabolism reactome fatty acid metabolism, arachidonic acid metabolism, vitamin B12 metabolism were positively enriched in DLBCL patients (Fig. 3C). More details of the GSEA enrichment can be seen in Supplementary Table S5. For validation of the relationships among 26 ER stress-related DEGs, the STRING tool was used to assess with confidence (value ≥ 0.40), and there were 26 nodes and 15 edges with PPI network enrichment (Fig. 4A). The top ER stress-related DEGs based on MCC score were identified based on the cytoHubba plugin in Cytoscape software. There are 9 hub genes, including TP53, CCL2, CEBPB, NUPR1, TRIB3, CAV1, UBE4B, NPLOC4, and NRIH3 (Fig. 4B), indicating they might play a significant role in DLBCL.

The functional enrichment analysis of ER stress-related DEGs. (A) GO enrichment analysis of ER stress-related DEGs. (B) KEGG pathway analysis of ER stress-related DEGs. (C) The enriched metabolic related pathways in DLBCL patients analyzed by GSEA. DEGs differentially expressed genes, ER endoplasmic reticulum, GO gene ontology, GSEA gene set enrichment analysis, KEGG kyoto encyclopedia of genes and genomes.

PPI network plot and hub genes of ER stress-related DEGs. (A) The PPI network of ER stress-related DEGs was visualized by Cytoscape software. (B) The hub genes were identified with the MCC score using cytoHubba plugin in Cytoscape software. PPI protein–protein interaction, ER endoplasmic reticulum, DEGs differentially expressed genes.
Identification of significant prognostic gene
To identify the significant ER stress-related genes and survival data in DLBCL patients, we analyzed the prognostic value of ER stress-related genes using the GEO datasets (GSE10846 and GSE11318) and the TCGA-DLBC dataset, which the clinical characteristics are shown in Table 1. It was found that NUPR1 and TRIB3 had survival differences in the above 3 datasets (Fig. 5A) and were among 26 ER stress-related DEGs. Interestingly, according to the previous PPI results, NUPR1 and TRIB3 genes might have a direct interaction, and the connection score was 0.439 (Fig. 5B). The KM curve showed that there was a significant difference between high and low expression of NUPR1 and TRIB3 (P < 0.01) in the GSE10846 and GSE11318 datasets (Fig. 5C,D). In the TCGA-DLBC dataset, we found that NUPR1 and TRIB3 not only have survival differences in over survival (OS), but also have survival differences in progress free survival (PFS), disease specific survival (DSS), and disease free interval (DFI), as shown in Fig. 5E,F. These results showed that patients with high expression of NUPR1 and low expression of TRIB3 might have a better prognosis.

Prognosis-related genes and survival plot. (A) Venn diagram showed that 2 overlapped co-expressed prognosis-related genes were screened based on Log-rank test in the GSE10846, GSE11318, and TCGA_DLBC datasets. (B) Protein Interaction of NUPR1 and TRIB3 Genes. (C) The KM curves showed the OS of patients in the high-risk and low-risk groups in GSE10846 dataset. (D) The KM curves showed the OS of patients in the high-risk and low-risk groups in GSE11318 dataset. (E) The up-regulation NUPR1 might be associated with better OS, PFS, DSS and DFI in TCGA_DLBC dataset. (F) The up-regulation TRIB3 might be associated with poor OS, PFS, DSS in TCGA_DLBC dataset. DSS disease specific survival; DFI disease free interval; KM Kaplan–Meier, OS over survival, PFS progression free survival, TCGA the cancer genome atlas.
Validation of the differential expression of NUPR1, TRIB3
We detected the expression of NUPR1 and TRIB3 in the GSE56315 dataset, the decreased expression of NUPR1 and increased expression of TRIB3 in DLBCL patients compared with NCs were showed, and the AUC was 0.791 and 0.807, respectively (Fig. 6A,B,D,E). Meanwhile, pan-cancer analysis based on the TIMER2 database41 (http://timer.cistrome.org) showed that NUPR1 had an inconsistent expression trend in different tumors, the expression of NUPR1 was decreased in bladder cancer, colon cancer, lung adenocarcinoma, prostate cancer, and other tumors. On the other hand, in breast cancer, renal clear cell carcinoma, and liver cancer, the expression of NUPR1 was increased (Fig. 6C). The upregulated TRIB3 might play an oncogenic role in tumors (Fig. 6F).

The identification of NUPR1 and TRIB3 in DLBCL. (A) Comparison of NUPR1 expression level of DLBCL and NCs in GSE56315 dataset. (B) The ROC curves of NUPR1 in GSE56315 dataset. (C) Pan-cancer analysis of NUPR1 in cancer tissues and normal counterparts from TCGA-DLBC databases. D. Comparison of TRIB3 expression level of DLBCL and NCs in GSE56315 dataset. (E) The ROC curves of TRIB3 in GSE56315 dataset. (F) Pan-cancer analysis of TRIB3 in TCGA databases. DLBCL diffuse large B cell lymphoma; ROC receiver operating characteristic; TCGA the cancer genome atlas.
Next, the correlation of NUPR1 and TRIB3 with the clinical characteristics of the GSE10864 and GSE11318 datasets was confirmed. The COO of DLBCL often identified three subgroups including activated B-cell-like (ABC), germinal-center B-cell-like (GCB), and unclassified (UNC). As shown in Fig. 7, The expression of NUPR1 in the ABC subtype was lower than in the GCB subtype both two GEO datasets (Fig. 7A,C), On the contrary, the expression of TRIB3 in the ABC subtype was higher than GCB subtype (Fig. 7E,G). However, the expression of NUPR1 and TRIB3 in differential stages was not significantly different based on the two GEO datasets (Fig. 7B,D,F,H). It showed that downregulated NUPR1 and upregulated TRIB3 might be identified as vital biomarkers for DLBCL.

Differential expression of NUPR1 and TRIB3. (A)–(B). The expression of NUPR1 among different subtypes and stages in GSE10846 dataset; (C)–(D). The expression of NUPR1 among different subtypes and stages in GSE11318 dataset; (E)–(F). The expression of TRIB3 among different subtypes and stages in GSE10846 dataset; (G)–(H). The expression of TRIB3 among different subtypes and stages in GSE11318 dataset; The qRT-PCR validation, (I) The expression of NUPR1 of GM12878, HBL-1, SUDHL2 and SUDHL4; (J) The expression of TRIB3 of GM12878, HBL-1, SUDHL2 and SUDHL4. * P < 0.05; ** P < 0.01; *** P < 0.001. qRT-PCR: quantitative real‐time polymerase chain reaction.
The expression of NUPR1 and TRIB3 in cultured cell lines
We also detected the expression of NUPR1 and TRIB3 in different cell lines via qRT-PCR (Fig. 7I,J). The expression in TRIB3 of DLBCL cell lines, including ABC subtype cell lines (HBL-1, SUDHL2) and GCB subtype cell lines (SUDHL4), were higher than normal B cell lines (GM12878) (P < 0.05), and ABC subtype cell lines were higher than GCB subtype (P < 0.05). But the expression of NUPR1 of HBL-1 had no significant difference in comparison with GM12878 (P = 0.085), the expression of NUPR1 in SUDHL2 and SUDHL4 was higher than GM12878 (P < 0.05). Both the expression of NUPR1 and TRIB3 in SUDHL2 than in SUDHL4 (P < 0.05).
Relationship between the expression of NUPR1, TRIB3 and immune infiltration
Based on our previous analysis, we speculated that NUPR1 might inhibit the progress of DLBCL and TRIB3 might promote it. Subsequently, the relationship between the expression of NUPR1, TRIB3, and immune infiltration was analyzed. As shown in Fig. 8, in the GSE10846 and GSE11318 datasets, NUPR1 was positively correlated with the estimate score, immune score, and matrix score, simultaneously, the expression of NUPR1 was positively correlated with M0 macrophages (Fig. 8A–D). However, TRIB3 was significantly negatively correlated with the estimate score, immune score, and matrix score (Fig. 8E–H), and had no significance with all immune cells in both two GEO datasets. These results suggested that NUPR1 might inhibit the progress of DLBCL by involving the immune response. But TRIB3 had no close relationship with immune cells through correlation with tumor purity.

The correlation of prognostic genes with immune infiltration. (A) The relationship between the expression of NUPR1 with ESTIMATE score, immune score and matrix score in the GSE10846 dataset. (B) The relationship between the expression of NUPR1 and the different subsets of immune cell infiltrates in the GSE10846 dataset. (C) The relationship between the expression of TRIB3 with ESTIMATE score, immune score and matrix score in the GSE10846 dataset. (D) The relationship between the expression of TRIB3 and the different subsets of immune cell infiltrates in the GSE10846 dataset. (E) The relationship between the expression of NUPR1 with ESTIMATE score, immune score and matrix score in the GSE11318 dataset. (F) The relationship between the expression of NUPR1 and the different subsets of immune cell infiltrates in the GSE11318 dataset. (G) The relationship between the expression of TRIB3 with ESTIMATE score, immune score and matrix score in the GSE11318 dataset. (H) The relationship between the expression of TRIB3 and the different subsets of immune cell infiltrates in the GSE11318 dataset. |rho|> 0.4 and P < 0.05 were considered to be significantly correlated.
Construction and validation of prognosis model
Based on the GSE10846 and GSE11318 datasets, a clinical prognosis model was developed using NUPR1 and TRIB3 as characteristic variables and its accuracy and generalizability assessed. By combining NUPR1 and TRIB3 with other clinical characteristics of DLBCL patients, such as gender, age, diagnosis type, stage, LDH ratio, number of extranodal lymph nodes, and ECOG score. We found that the above risk factors were statistically different in univariate analysis except for gender. Subsequently, multivariate Cox analysis found that age (≥ 60 years old), stage, LDH ratio, and the expression of TRIB3 were independent prognostic factors (Fig. 9A). Next, the nomogram for accurate patient prognosis and prediction was constructed based on the clinical characteristics of DLBCL patients and the expression of TRIB3 (Fig. 9B). In order to evaluate the accuracy of the model, the time-dependent ROC curve was drawn using the GSE10846 dataset as the training cohort and the GSE11318 dataset as the test cohort. The AUC corresponding to 1, 3, and 5 years of both the training cohort and test cohort were all above 0.75 (Fig. 9C–D), indicating the nomogram model provided a good predictive accuracy of DLBCL. Furthermore, the calibration curve suggested that the consistency of the model is good at 1-, 3-, and 5-year intervals (Fig. 9E). These above results indicated that combining the clinical characteristics of DLBCL with the expression of TRIB3 could predict the prognosis of patients with DLBCL in 1-, 3-, and 5-year intervals, which was expected to provide a more effective reference for clinicians and formulate effective intervention measures.

Construction and validation of prognosis model. (A) Forest plot summary of the univariate and multivariable analyses of NUPR1, TRIB3 and other clinical characteristics. (B) Nomogram for predicting the probability of patient mortality at 1-, 3- and 5-year of OS. (C) The ROC for 1-, 2- and 3-year survival rate in the GSE10846 dataset, and the AUC was 0.785, 0.786 and 0.792, respectively. (D) The ROC for 1-, 2- and 3-year survival rate in the GSE11318 dataset, and the AUC was 0.754, 0.781 and 0.808, respectively. (E) 1-, 3-, and 5-year calibration curves of clinical prediction models for DLBCL patients, the model is validated by resampling with the bootstrap method, and the number of times was 1000. AUC area under curve, DLBCL diffuse large B cell lymphoma, OS overall survival.
Discussion
Due to differences in gene expression profiles and genetic alterations, DLBCL is a highly diverse lymphoid neoplasm that exhibits a wide range of clinical outcomes and therapeutic responses2,3,9. There has been significant development in targeted treatment and immunotherapy for DLBCL in recent years3,5, however, innovative treatment approaches and targets are urgently required. ER stress has emerged as a focal-point and forward position field in a variety of human malignancies during the last decade22,23, and is involved in many biological processes, such as apoptosis, autophagy42, ferroptosis43, and hypoxia44. But there is a lack of in-depth knowledge about the role of ER stress in the clinical progress of DLBCL.
So far as we know, our study is the first research on the relationship between the ER stress-related gene and DLBCL. These findings should help researchers in the future learn more about how to predict prognosis and treat DLBCL patients individually in the clinic. Based on the outcomes of previous bioinformatic analysis, we have found nine genes may play essential roles in DLBCL. As previously described45,46,47, it had been proven that TP53, CCL2, and CEBPB were involved in the carcinogenesis and development of DLBCL. TP53 has been identified as one of the most frequently mutated genes and could be a valuable prognostic biomarker in both ABC and GCB DLBCL patients45,46. It was found that CEBPB and BCL2A1 can induce cell transformation and increase the survival of anaplastic large cell lymphomas cells47. However, no research has been reported about the role of NUPR1, TRIB3, CAV1, UBE4B, NPLOC4, and NRIH3 in DLBCL. After identifying the prognostic value of ER stress-related genes using three datasets, NUPR1 and TRIB3 may play important roles in DLBCL carcinogenesis and development. NUPR1 is located in the nucleus of various cells, including cancer cells48,49. Borrello et al. demonstrated the essential role of NUPR1 which participated in the regulation of UPR and more broadly in the integrated stress response by interacting with eIF2α, and protected the liver from metabolic distress by controlling lipid homeostasis50. Howerer, Liu et al. found that NUPR1 was upregulated in the bone marrow of patients with multiple myeloma (MM)51, downregulation of NUPR1 might significantly inhibit cell proliferation and promote autophagy-mediated apoptosis in MM52. NUPR1 could also active autophagy and bind to the promoter regions of some autophagy-related genes, such as BECN1, GREB1, RAB31, PGR, CYP1B1, thereby regulating breast cancer metastasis and transcription49,53. Histone methyltransferase Dot1L might inhibit pancreatic cancer cell apoptosis by targeting NUPR1, and overexpressed NUPR1 also inhibited pancreatic cancer cell apoptosis54. In summary, NUPR1 can play an important role in cell stress and stress‑related apoptosis48,55. We found the level of NUPR1 in DLBCL patients to be downregulated, and that in the GCB subtype was significantly higher than the ABC subtype in our study. We also found NUPR1 could involve in the immune response.
Compared with noncancerous tissues, TRIB3 expression was markedly increased in DLBCL patients, particularly in the ABC subtype and later stages, suggesting that TRIB3 may play a carcinogenic role in DLBCL56,57,58,59. As a member of the mammalian pseudokinase tribbles family, TRIB3 can interfere with a lot of proteins, such as kinase-dependent proteins, transcription factors, and ubiquitin ligases60,61. Evidences show that TRIB3 can regulate the downstream biological process of ER stress, alleviate cell stress, and promote cell survival58,62,63. Ohoka et al. demonstrated that certain ER stress inducers, including low glucose and hypoxia, may increase TRIB3 expression62,63,64. Twist Family BHLH Transcription Factor 1 (TWIST1) is stabilized in part by TRIB3 binding to its WR domain and blocking its ubiquitination, depletion of TRIB3 can boost TWIST1 degradation and increase sensitivity to all-trans retinoic acid (ATRA), consistent with its role in regulating carcinogenesis and progression63. TRIB3 overexpression in human gastric cancer was related to tumor angiogenesis and a poor prognosis65. Through analysis of lymphoma specimens, TRIB3 expression was positively correlated with MYC expression66, MYC is a transcription factor, and its alterations have been considered to be associated with aggressive clinical behavior in DLBCL67. Mechanistically, E3 ubiquitin ligase UBE3B-mediated MYC ubiquitination and degradation can be inhibited by TRIB3 bind to MYC66. These results are in agreement with our studies and imply that TRIB3 may be a critical regulatory factor in driving cancer cell proliferation, migration, and invasion57,58,68. Furthermore, our bioinformatic analysis indicated that TRIB3 could be a significant survival predictor and a potential therapeutic target for DLBCL.
Our bioinformatic analysis suggested a direct connection between NUPR1 and TRIB3. Methamphetamine (METH) exposed to rat and PC12 cell lines could increase the expression of NUPR1 accompanied by CHOP and TRIB3 upregulation and promote apoptosis and autophagy69, suggesting the NUPR1/CHOP/TRIB3 signal pathway plays a crucial function in the regulation of apoptosis and autophagy via ER stress69,70. There were shown the expression of NUPR1 and TRIB3 concurrently. Unfortunately, our study suggested that both the lower expression of NUPR1 and the higher expression of TRIB3 meant a worse clinical prognosis for DLBCL patients. Furthermore, the qRT-PCR test showed the expression of TRIB3 in HBL-1, SUDHL2, and SUDHL4 was higher than GM12878, and these results were consistent with our bioinformatic analysis, However, the expression of NUPR1 was not compatible with these analysis.
The clinical prognostic model was built based on the GSE10846 and GSE11318 datasets, using NUPR1 and TRIB3 as feature variables, and evaluated for accuracy and generalizability. There are some authors71,72 who have built the prognosis model that is similar to ours. Lv et al. showed that carcinoembryonic antigen, N stage, and surgical method were independent prognostic factors for overall survival in patients with obstructive colorectal cancer using a nomogram model71. For researching the role of ACK1-associated immunomodulators in non-small cell lung cancer (NSCLC), Zhu J et al. Established a multiple-gene risk prediction model, and the results showed the risk scores were an independent prognosis predictor in the TCGA lung cohorts72. It was suggested that ACK1 may be a potential immunotherapeutic target72. Numerous types of tumors may share similar genetic or molecular features, there are a lot of studies48,55,57,60,61 showed that NUPR1 and TRIB3 are involved in cancer biology, and we think this prognosis model that includes these genes may be applied to more types of tumors. However, due to the specificity and complexity of different types of cancer, further studies should verify and adjust the accuracy and reliability of these identical markers and explore the relationship between their biological functions and clinical applications in numerous types of tumors.
Some limitations existed in this study. First, the prognostic model was built and validated using publicly available data; hence, more prospective studies are needed to confirm its accuracy and utility. Second, by narrowing our focus to only two ER stress-related genes, NUPR1 and TRIB3, we may have missed other noteworthy prognostic genes for DLBCL. Third, only four cell lines were used to verify the expression of NUPR1 and TRIB3, and we will carry out more cell lines for research if the experimental conditions are confirmed.
Conclusion
In summary, our study clarified that downregulation the expression of NUPR1 and upregulation of TRIB3 in DLBCL patients. Further study has shown NUPR1 may inhibit the progress of DLBCL by involving the immune response, and TRIB3 should be a carcinogenic gene in DLBCL through regulating ER stress. Moreover, we found that combining the clinical characteristics of DLBCL by TRIB3 expression could better predict the prognosis of DLBCL patients, suggesting TRIB3 might serve as a potential therapeutic target and prognostic factor in DLBCL.
Data availability
All of datasets used in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
References
-
Swerdlow, S. H. et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127, 2375–2390 (2016).
Google Scholar
-
Sehn, L. H. & Salles, G. Diffuse large B-cell lymphoma. N. Engl. J. Med. 384, 842–858 (2021).
Google Scholar
-
Sarkozy, C. & Sehn, L. H. Management of relapsed/refractory DLBCL. Best Pract. Res. Clin. Haematol. 31, 209–216 (2018).
Google Scholar
-
Coiffier, B. et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N. Engl. J. Med. 346, 235–242 (2002).
Google Scholar
-
Lugtenburg, P. J. et al. Rituximab-CHOP with early rituximab intensification for diffuse large B-cell lymphoma: A randomized phase III trial of the HOVON and the nordic lymphoma group (HOVON-84). J. Clin. Oncol. 38, 3377–3387 (2020).
Google Scholar
-
Vaidya, R. & Witzig, T. E. Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Ann. Oncol. 25, 2124–2133 (2014).
Google Scholar
-
Kondo, E. Autologous hematopoietic stem cell transplantation for diffuse large B-cell lymphoma. Anal. Real-World Data in Patients Relapsed/Refract. Diffuse Large B Cell Lymphoma Who Receiv. Salvage Chemother. Rituximab Era 56, 100–108 (2016).
-
Fuji, S. et al. Analysis of real-world data in patients with relapsed/refractory diffuse large B cell lymphoma who received salvage chemotherapy in the rituximab era. Ann. Hematol. 100, 2253–2260 (2021).
Google Scholar
-
Miao, X. et al. Mitochondrial dysfunction in endothelial cells induced by airborne fine particulate matter (2.5 m). J. Appl. Toxicol. 39, 1424–1432 (2019).
Google Scholar
-
Xu-Monette, Z. Y. et al. A refined cell-of-origin classifier with targeted NGS and artificial intelligence shows robust predictive value in DLBCL. Blood Adv. 4, 3391–3404 (2020).
Google Scholar
-
Rosenwald, A. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346, 1937–1947 (2002).
Google Scholar
-
Wight, J. C. et al. Prognostication of diffuse large B-cell lymphoma in the molecular era: Moving beyond the IPI. Blood Rev. 32, 400–415 (2018).
Google Scholar
-
Wang, H.-T. et al. Acrolein induces mtDNA damages, mitochondrial fission and mitophagy in human lung cells. Oncotarget 8, 70406–70421 (2017).
Google Scholar
-
Su, W. et al. A novel classification based on B-cell receptor signal gene expression correlates with prognosis in primary breast diffuse large B-cell lymphoma. J. Cancer 11, 2431–2441 (2020).
Google Scholar
-
Liu, X. et al. Mitochondrial-endoplasmic reticulum communication-mediated oxidative stress and autophagy. Biomed. Res. Int. 2022, 6459585 (2022).
Google Scholar
-
Ye, J. & Liu, X. Interactions between endoplasmic reticulum stress and extracellular vesicles in multiple diseases. Front. Immunol. 13, 955419 (2022).
Google Scholar
-
Cui, Y. et al. The regulation of miR-320a/XBP1 axis through LINC00963 for endoplasmic reticulum stress and autophagy in diffuse large B-cell lymphoma. Cancer Cell. Int. 21, 305 (2021).
Google Scholar
-
Mahadevan, N. R. et al. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc. Natl. Acad. Sci. U. S. A. 108, 6561–6566 (2011).
Google Scholar
-
Senft, D. & Ronai, Z. A. UPR, autophagy and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 40, 141–148 (2015).
Google Scholar
-
Tian, X. et al. Targeting the integrated stress response in cancer therapy. Front. Pharmacol. 12, 747837 (2021).
Google Scholar
-
Lin, Y.-Z. et al. Roundabout signaling pathway involved in the pathogenesis of COPD by integrative bioinformatics analysis. Int. J. Chron. Obstruct. Pulmon. Dis. 14, 2145–2162 (2019).
Google Scholar
-
Kumar, V. & Maity, S. ER stress-sensor proteins and ER-mitochondrial crosstalk (ER) stress response. Biomolecules 11, 173 (2021).
Google Scholar
-
Oakes, S. A. & Papa, F. R. The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 10, 173–194 (2015).
Google Scholar
-
Dybkær, K. et al. Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J. Clin. Oncol. 33, 1379–1388 (2015).
Google Scholar
-
Lenz, G. et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 359, 2313–2323 (2008).
Google Scholar
-
Lenz, G. et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. U. S. A. 105, 13520–13525 (2008).
Google Scholar
-
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Google Scholar
-
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Google Scholar
-
Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res 43, D1049-1056 (2015).
Google Scholar
-
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27, 29–34 (1999).
Google Scholar
-
Yu, G. et al. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Google Scholar
-
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 (2005).
Google Scholar
-
Szklarczyk, D. et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368 (2017).
Google Scholar
-
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Google Scholar
-
Chin, C.-H. et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8(Suppl 4), S11 (2014).
Google Scholar
-
Martínez-Camblor, P. et al. Efficient nonparametric confidence bands for receiver operating-characteristic curves. Stat. Methods Med. Res. 27, 1892–1908 (2018).
Google Scholar
-
Rusk, N. Expanded CIBERSORTx. Nat. Methods 16, 577 (2019).
Google Scholar
-
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Google Scholar
-
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Google Scholar
-
Davis, S. & Meltzer, P. S. GEOquery: A bridge between the gene expression omnibus (GEO) and bioconductor. Bioinformatics 23, 1846–1847 (2007).
Google Scholar
-
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48, W509–W514 (2020).
Google Scholar
-
Qi, Z. & Chen, L. Endoplasmic reticulum stress and autophagy. Adv. Exp. Med. Biol. 1206, 167–177 (2019).
Google Scholar
-
Lee, Y.-S. et al. Ferroptosis-induced endoplasmic reticulum stress: Cross-talk between ferroptosis and apoptosis. Mol. Cancer Res. 16, 1073–1076 (2018).
Google Scholar
-
Akman, M. et al. Hypoxia, endoplasmic reticulum stress and chemoresistance: Dangerous liaisons. J. Exp. Clin. Cancer Res. 40, 28 (2021).
Google Scholar
-
Qin, Y. et al. Characteristics and management of TP53-mutated diffuse large B-cell lymphoma patients. Cancer Manag. Res. 12, 11515–11522 (2020).
Google Scholar
-
Voropaeva, E. N. et al. Clinical aspects of TP53 gene inactivation in diffuse large B-cell lymphoma. BMC Med. Genom. 12, 35 (2019).
Google Scholar
-
Piva, R. et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J. Clin. Invest. 116, 3171–3182 (2006).
Google Scholar
-
Martin, T. A. et al. NUPR1 and its potential role in cancer and pathological conditions (review). Int. J. Oncol. 58, 21 (2021).
Google Scholar
-
Xiao, H. et al. NUPR1 promotes the proliferation and migration of breast cancer cells by activating TFE3 transcription to induce autophagy. Exp. Cell Res. 418, 113234 (2022).
Google Scholar
-
Borrello, M. T. et al. NUPR1 interacts with eIF2 α and is required for resolution of the ER stress response in pancreatic tissue. FEBS J. 288, 4081–4097 (2021).
Google Scholar
-
Liu, J.-Y. et al. The underlying role of mitophagy in different regulatory mechanisms of chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 15, 2167–2177 (2020).
Google Scholar
-
Chen, X. et al. microRNA-637 promotes apoptosis and suppresses proliferation and autophagy in multiple myeloma cell lines via NUPR1. FEBS Open Bio 11, 519–528 (2021).
Google Scholar
-
Wang, L. et al. Transcriptional coregualtor NUPR1 maintains tamoxifen resistance in breast cancer cells. Cell Death Dis. 12, 149 (2021).
Google Scholar
-
Shan, L. et al. Histone methyltransferase Dot1L inhibits pancreatic cancer cell apoptosis by promoting NUPR1 expression. J. Int. Med. Res. 50, 3000605221088431 (2022).
Google Scholar
-
Chowdhury, U. R. et al. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 28, 225–232 (2009).
Google Scholar
-
Teresa Borrello, M. et al. NUPR1 protects liver from lipotoxic injury by improving the endoplasmic reticulum stress response. FASEB J. 35, e21395 (2021).
Google Scholar
-
Sakai, S. et al. Tribbles-related protein family members as regulators or substrates of the ubiquitin-proteasome system in cancer development. Curr. Cancer Drug Targets 16, 147–156 (2016).
Google Scholar
-
Chen, Q.-Z. et al. TRIB3 interacts with STAT3 to promote cancer angiogenesis. Curr. Med. Sci. 42, 932–940 (2022).
Google Scholar
-
Hernández-Quiles, M. et al. TRIB3 modulates PPARγ-mediated growth inhibition by interfering with the MLL complex in breast cancer cells. Int. J. Mol. Sci. 23, 10535 (2022).
Google Scholar
-
Stefanovska, B. et al. Tribbles Pseudokinase 3 regulation and contribution to cancer. Cancers (Basel) 13, 1822 (2021).
Google Scholar
-
Mondal, D. et al. Tripping on TRIB3 at the junction of health, metabolic dysfunction and cancer. Biochimie 124, 34–52 (2016).
Google Scholar
-
Ohoka, N. et al. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24, 1243–1255 (2005).
Google Scholar
-
Lin, J. et al. TRIB3 stabilizes high TWIST1 expression to promote rapid APL progression and ATRA resistance. Clin. Cancer Res. 25, 6228–6242 (2019).
Google Scholar
-
Zhu, Y. et al. Sp2 promotes invasion and metastasis of hepatocellular carcinoma by targeting TRIB3 protein. Cancer Med. 9, 3592–3603 (2020).
Google Scholar
-
Dong, S. et al. Overexpression of TRIB3 promotes angiogenesis in human gastric cancer. Oncol. Rep. 36, 2339–2348 (2016).
Google Scholar
-
Li, K. et al. TRIB3 promotes MYC-associated lymphoma development through suppression of UBE3B-mediated MYC degradation. Nat. Commun. 11, 6316 (2020).
Google Scholar
-
Ott, G. et al. Understanding MYC-driven aggressive B-cell lymphomas: Pathogenesis and classification. Blood 122, 3884–3891 (2013).
Google Scholar
-
Orea-Soufi, A. et al. The Pseudokinase TRIB3 negatively regulates the HER2 receptor pathway and is a biomarker of good prognosis in luminal breast cancer. Cancers (Basel) 13, 5307 (2021).
Google Scholar
-
Xu, X. et al. Nupr1 modulates methamphetamine-induced dopaminergic neuronal apoptosis and autophagy through CHOP-Trib3-mediated endoplasmic reticulum stress signaling pathway. Front Mol. Neurosci. 10, 203 (2017).
Google Scholar
-
Tabatabaei Mirakabad, F. S. et al. NUPR1- CHOP experssion, autophagosome formation and apoptosis in the postmortem striatum of chronic methamphetamine user. J. Chem. Neuroanat. 114, 101942 (2021).
Google Scholar
-
Lv, J. et al. A nomogram model for predicting prognosis of obstructive colorectal cancer. World J. Surg. Oncol. 19, 337 (2021).
Google Scholar
-
Zhu, J. et al. Comprehensive analysis of the immune implication of ACK1 gene in non-small cell lung cancer. Front Oncol. 10, 1132 (2020).
Google Scholar
Acknowledgements
This study was supported by the General Program of the Natural Science Foundation of Fujian Province, China (No. 2022J011435, 2020J011251).
Author information
Authors and Affiliations
Contributions
Z.C., L.Q., and Z.X. designed and directed the project; Z.C., L.C., D.M., and W.H. derived the models and analysed the data; Z.C. and L.Q. wrote the manuscript; Z.X. supervised the entire study; Z.X. and C.Z. reviewed and edited the manuscript. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Tables.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Zhang, C., Lin, Q., Li, C. et al. Analysis of endoplasmic reticulum stress-related gene signature for the prognosis and pattern in diffuse large B cell lymphoma.
Sci Rep 13, 13894 (2023). https://doi.org/10.1038/s41598-023-38568-x
-
Received: 03 April 2023
-
Accepted: 11 July 2023
-
Published: 25 August 2023
-
DOI: https://doi.org/10.1038/s41598-023-38568-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.