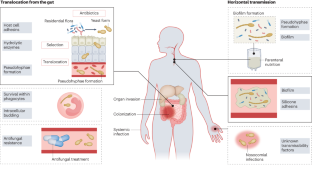
Infection
Candida parapsilosis complex in the clinical setting
Abstract
Representatives of the Candida parapsilosis complex are important yeast species causing human infections, including candidaemia as one of the leading diseases. This complex comprises C. parapsilosis, Candida orthopsilosis and Candida metapsilosis, and causes a wide range of clinical presentations from colonization to superficial and disseminated infections with a high prevalence in preterm-born infants and the potential to cause outbreaks in hospital settings. Compared with other Candida species, the C. parapsilosis complex shows high minimal inhibitory concentrations for echinocandin drugs due to a naturally occurring FKS1 polymorphism. The emergence of clonal outbreaks of strains with resistance to commonly used antifungals, such as fluconazole, is causing concern. In this Review, we present the latest medical data covering epidemiology, diagnosis, resistance and current treatment approaches for the C. parapsilosis complex. We describe its main clinical manifestations in adults and children and highlight new treatment options. We compare the three sister species, examining key elements of microbiology and clinical characteristics, including the population at risk, disease manifestation and colonization status. Finally, we provide a comprehensive resource for clinicians and researchers focusing on Candida species infections and the C. parapsilosis complex, aiming to bridge the emerging translational knowledge and future therapeutic challenges associated with this human pathogen.
This is a preview of subscription content, access via your institution
Access options
style{display:none!important}.LiveAreaSection-193358632 *{align-content:stretch;align-items:stretch;align-self:auto;animation-delay:0s;animation-direction:normal;animation-duration:0s;animation-fill-mode:none;animation-iteration-count:1;animation-name:none;animation-play-state:running;animation-timing-function:ease;azimuth:center;backface-visibility:visible;background-attachment:scroll;background-blend-mode:normal;background-clip:borderBox;background-color:transparent;background-image:none;background-origin:paddingBox;background-position:0 0;background-repeat:repeat;background-size:auto auto;block-size:auto;border-block-end-color:currentcolor;border-block-end-style:none;border-block-end-width:medium;border-block-start-color:currentcolor;border-block-start-style:none;border-block-start-width:medium;border-bottom-color:currentcolor;border-bottom-left-radius:0;border-bottom-right-radius:0;border-bottom-style:none;border-bottom-width:medium;border-collapse:separate;border-image-outset:0s;border-image-repeat:stretch;border-image-slice:100%;border-image-source:none;border-image-width:1;border-inline-end-color:currentcolor;border-inline-end-style:none;border-inline-end-width:medium;border-inline-start-color:currentcolor;border-inline-start-style:none;border-inline-start-width:medium;border-left-color:currentcolor;border-left-style:none;border-left-width:medium;border-right-color:currentcolor;border-right-style:none;border-right-width:medium;border-spacing:0;border-top-color:currentcolor;border-top-left-radius:0;border-top-right-radius:0;border-top-style:none;border-top-width:medium;bottom:auto;box-decoration-break:slice;box-shadow:none;box-sizing:border-box;break-after:auto;break-before:auto;break-inside:auto;caption-side:top;caret-color:auto;clear:none;clip:auto;clip-path:none;color:initial;column-count:auto;column-fill:balance;column-gap:normal;column-rule-color:currentcolor;column-rule-style:none;column-rule-width:medium;column-span:none;column-width:auto;content:normal;counter-increment:none;counter-reset:none;cursor:auto;display:inline;empty-cells:show;filter:none;flex-basis:auto;flex-direction:row;flex-grow:0;flex-shrink:1;flex-wrap:nowrap;float:none;font-family:initial;font-feature-settings:normal;font-kerning:auto;font-language-override:normal;font-size:medium;font-size-adjust:none;font-stretch:normal;font-style:normal;font-synthesis:weight style;font-variant:normal;font-variant-alternates:normal;font-variant-caps:normal;font-variant-east-asian:normal;font-variant-ligatures:normal;font-variant-numeric:normal;font-variant-position:normal;font-weight:400;grid-auto-columns:auto;grid-auto-flow:row;grid-auto-rows:auto;grid-column-end:auto;grid-column-gap:0;grid-column-start:auto;grid-row-end:auto;grid-row-gap:0;grid-row-start:auto;grid-template-areas:none;grid-template-columns:none;grid-template-rows:none;height:auto;hyphens:manual;image-orientation:0deg;image-rendering:auto;image-resolution:1dppx;ime-mode:auto;inline-size:auto;isolation:auto;justify-content:flexStart;left:auto;letter-spacing:normal;line-break:auto;line-height:normal;list-style-image:none;list-style-position:outside;list-style-type:disc;margin-block-end:0;margin-block-start:0;margin-bottom:0;margin-inline-end:0;margin-inline-start:0;margin-left:0;margin-right:0;margin-top:0;mask-clip:borderBox;mask-composite:add;mask-image:none;mask-mode:matchSource;mask-origin:borderBox;mask-position:0 0;mask-repeat:repeat;mask-size:auto;mask-type:luminance;max-height:none;max-width:none;min-block-size:0;min-height:0;min-inline-size:0;min-width:0;mix-blend-mode:normal;object-fit:fill;object-position:50% 50%;offset-block-end:auto;offset-block-start:auto;offset-inline-end:auto;offset-inline-start:auto;opacity:1;order:0;orphans:2;outline-color:initial;outline-offset:0;outline-style:none;outline-width:medium;overflow:visible;overflow-wrap:normal;overflow-x:visible;overflow-y:visible;padding-block-end:0;padding-block-start:0;padding-bottom:0;padding-inline-end:0;padding-inline-start:0;padding-left:0;padding-right:0;padding-top:0;page-break-after:auto;page-break-before:auto;page-break-inside:auto;perspective:none;perspective-origin:50% 50%;pointer-events:auto;position:static;quotes:initial;resize:none;right:auto;ruby-align:spaceAround;ruby-merge:separate;ruby-position:over;scroll-behavior:auto;scroll-snap-coordinate:none;scroll-snap-destination:0 0;scroll-snap-points-x:none;scroll-snap-points-y:none;scroll-snap-type:none;shape-image-threshold:0;shape-margin:0;shape-outside:none;tab-size:8;table-layout:auto;text-align:initial;text-align-last:auto;text-combine-upright:none;text-decoration-color:currentcolor;text-decoration-line:none;text-decoration-style:solid;text-emphasis-color:currentcolor;text-emphasis-position:over right;text-emphasis-style:none;text-indent:0;text-justify:auto;text-orientation:mixed;text-overflow:clip;text-rendering:auto;text-shadow:none;text-transform:none;text-underline-position:auto;top:auto;touch-action:auto;transform:none;transform-box:borderBox;transform-origin:50% 50%0;transform-style:flat;transition-delay:0s;transition-duration:0s;transition-property:all;transition-timing-function:ease;vertical-align:baseline;visibility:visible;white-space:normal;widows:2;width:auto;will-change:auto;word-break:normal;word-spacing:normal;word-wrap:normal;writing-mode:horizontalTb;z-index:auto;-webkit-appearance:none;-moz-appearance:none;-ms-appearance:none;appearance:none;margin:0}.LiveAreaSection-193358632{width:100%}.LiveAreaSection-193358632 .login-option-buybox{display:block;width:100%;font-size:17px;line-height:30px;color:#222;padding-top:30px;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-access-options{display:block;font-weight:700;font-size:17px;line-height:30px;color:#222;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-login>li:not(:first-child)::before{transform:translateY(-50%);content:””;height:1rem;position:absolute;top:50%;left:0;border-left:2px solid #999}.LiveAreaSection-193358632 .additional-login>li:not(:first-child){padding-left:10px}.LiveAreaSection-193358632 .additional-login>li{display:inline-block;position:relative;vertical-align:middle;padding-right:10px}.BuyBoxSection-683559780{display:flex;flex-wrap:wrap;flex:1;flex-direction:row-reverse;margin:-30px -15px 0}.BuyBoxSection-683559780 .box-inner{width:100%;height:100%}.BuyBoxSection-683559780 .readcube-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:1;flex-basis:255px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:300px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox-nature-plus{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:100%;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .title-readcube,.BuyBoxSection-683559780 .title-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .title-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .asia-link{color:#069;cursor:pointer;text-decoration:none;font-size:1.05em;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:1.05em6}.BuyBoxSection-683559780 .access-readcube{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;opacity:.8px;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .price-buybox{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;padding-top:30px;text-align:center}.BuyBoxSection-683559780 .price-buybox-to{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center}.BuyBoxSection-683559780 .price-info-text{font-size:16px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-value{font-size:30px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-per-period{font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-from{font-size:14px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .issue-buybox{display:block;font-size:13px;text-align:center;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:19px}.BuyBoxSection-683559780 .no-price-buybox{display:block;font-size:13px;line-height:18px;text-align:center;padding-right:10%;padding-left:10%;padding-bottom:20px;padding-top:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .vat-buybox{display:block;margin-top:5px;margin-right:20%;margin-left:20%;font-size:11px;color:#222;padding-top:10px;padding-bottom:15px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:17px}.BuyBoxSection-683559780 .tax-buybox{display:block;width:100%;color:#222;padding:20px 16px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:NaNpx}.BuyBoxSection-683559780 .button-container{display:flex;padding-right:20px;padding-left:20px;justify-content:center}.BuyBoxSection-683559780 .button-container>*{flex:1px}.BuyBoxSection-683559780 .button-container>a:hover,.Button-505204839:hover,.Button-1078489254:hover,.Button-2496381730:hover{text-decoration:none}.BuyBoxSection-683559780 .readcube-button{background:#fff;margin-top:30px}.BuyBoxSection-683559780 .button-asia{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;margin-top:75px}.BuyBoxSection-683559780 .button-label-asia,.ButtonLabel-3869432492,.ButtonLabel-3296148077,.ButtonLabel-1651148777{display:block;color:#fff;font-size:17px;line-height:20px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center;text-decoration:none;cursor:pointer}.Button-505204839,.Button-1078489254,.Button-2496381730{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;max-width:320px;margin-top:10px}.Button-505204839 .readcube-label,.Button-1078489254 .readcube-label,.Button-2496381730 .readcube-label{color:#069}
/* style specs end */
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout


References
-
Brown, G. D. et al. Hidden killers: human fungal infections. Sci. Transl Med. 4, 165rv113 (2012).
-
Hawksworth, D. L. & Lucking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.funk-0052-2016 (2017).
-
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L. & Kullberg, B. J. Invasive candidiasis. Nat. Rev. Dis. Primers 4, 18026 (2018).
-
Rolling, T. et al. Haematopoietic cell transplantation outcomes are linked to intestinal mycobiota dynamics and an expansion of Candida parapsilosis complex species. Nat. Microbiol. 6, 1505–1515 (2021).
Google Scholar
-
Zhai, B. et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat. Med. 26, 59–64 (2020).
Google Scholar
-
Hoenigl, M. et al. COVID-19-associated fungal infections. Nat. Microbiol. 7, 1127–1140 (2022). This article provides an update on the epidemiology, clinical risk factors and predisposing features of the host environment of COVID-19-associated fungal infections.
Google Scholar
-
Ramos-Martínez, A. et al. Impact of the COVID-19 pandemic on the clinical profile of candidemia and the incidence of fungemia due to fluconazole-resistant Candida parapsilosis. J. Fungi 8, 451 (2022).
-
Prattes, J. et al. Diagnosis and treatment of COVID-19 associated pulmonary apergillosis in critically ill patients: results from a European confederation of medical mycology registry. Intensive Care Med. 47, 1158–1160 (2021).
Google Scholar
-
Krishna, V., Bansal, N., Morjaria, J. & Kaul, S. COVID-19-associated pulmonary mucormycosis. J. Fungi 8, 711 (2022).
Google Scholar
-
Asadzadeh, M. et al. High-resolution fingerprinting of Candida parapsilosis isolates suggests persistence and transmission of infections among neonatal intensive care unit patients in Kuwait. Sci. Rep. 9, 1340 (2019).
Google Scholar
-
Mamali, V. et al. Increasing incidence and shifting epidemiology of candidemia in Greece: results from the first nationwide 10-year survey. J. Fungi 8, 116 (2022).
Google Scholar
-
Arastehfar, A. et al. Candidemia among coronavirus disease 2019 patients in Turkey admitted to intensive care units: a retrospective multicenter study. Open Forum Infect. Dis. 9, ofac978 (2022).
-
Pfaller, M. A., Diekema, D. J., Turnidge, J. D., Castanheira, M. & Jones, R. N. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open Forum Infect. Dis. 6, S79–S94 (2019).
Google Scholar
-
Pfaller, M. A., Carvalhaes, C. G., DeVries, S., Rhomberg, P. R. & Castanheira, M. Impact of COVID-19 on the antifungal susceptibility profiles of isolates collected in a global surveillance program that monitors invasive fungal infections. Med. Mycol. 60, myac028 (2022).
Google Scholar
-
Diaz-Garcia, J. et al. Evidence of fluconazole-resistant Candida parapsilosis genotypes spreading across hospitals located in Madrid, Spain and harboring the Y132F ERG11p substitution. Antimicrob. Agents Chemother. 66, e0071022 (2022).
-
Toth, R. et al. Candida parapsilosis: from genes to the bedside. Clin. Microbiol. Rev. 32, e00111-18 (2019). This review discusses C. parapsilosis pathogenesis, transcriptome studies and molecular mechanisms of virulence.
Google Scholar
-
Nemeth, T. M. G., Gacser, A., Nosanchuk, J. D. A. in Encyclopedia of Mycology Vol. 1 526–543 (Elsevier, 2021).
-
Mixao, V. et al. Genome analysis of five recently described species of the CUG-Ser clade uncovers Candida theae as a new hybrid lineage with pathogenic potential in the Candida parapsilosis species complex. DNA Res. 29, dsac010 (2022).
Google Scholar
-
Samantaray, S. & Singh, R. Evaluation of MALDI-TOF MS for identification of species in the Candida parapsilosis complex from candidiasis cases. J. Appl. Lab. Med. 7, 889–900 (2022).
-
Krassowski, T. et al. Evolutionary instability of CUG-Leu in the genetic code of budding yeasts. Nat. Commun. 9, 1887 (2018).
Google Scholar
-
Pountain, A. W., Collette, J. R., Farrell, W. M. & Lorenz, M. C. Interactions of both pathogenic and nonpathogenic CUG clade Candida species with macrophages share a conserved transcriptional landscape. mBio 12, e0331721 (2021).
-
Gómez-Molero, E. et al. Phenotypic variability in a coinfection with three independent Candida parapsilosis lineages. Front. Microbiol. 11, 1994 (2020).
Google Scholar
-
Pal, S. E. et al. A Candida parapsilosis overexpression collection reveals genes required for pathogenesis. J. Fungi 7, 97 (2021).
Google Scholar
-
Kulig, K. et al. Insight into the properties and immunoregulatory effect of extracellular vesicles produced by Candida glabrata, Candida parapsilosis, and Candida tropicalis biofilms. Front. Cell Infect. Microbiol. 12, 879237 (2022).
Google Scholar
-
Yamin, D. H., Husin, A. & Harun, A. Risk factors of Candida parapsilosis catheter-related bloodstream infection. Front. Public Health 9, 631865 (2021).
Google Scholar
-
Demirci-Duarte, S., Arikan-Akdagli, S. & Gulmez, D. Species distribution, azole resistance and related molecular mechanisms in invasive Candida parapsilosis complex isolates: increase in fluconazole resistance in 21 years. Mycoses 64, 823–830 (2021).
Google Scholar
-
Thomaz, D. Y. et al. Environmental clonal spread of azole-resistant Candida parapsilosis with Erg11-Y132F mutation causing a large candidemia outbreak in a Brazilian cancer referral center. J. Fungi 7, 259 (2021).
Google Scholar
-
Arastehfar, A. et al. Clonal candidemia outbreak by Candida parapsilosis carrying Y132F in Turkey: evolution of a persisting challenge. Front. Cell Infect. Microbiol. 11, 676177 (2021).
Google Scholar
-
Thomaz, D. Y. et al. An azole-resistant Candida parapsilosis outbreak: clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front. Microbiol. 9, 2997 (2018).
Google Scholar
-
Daneshnia, F. et al. Determinants of fluconazole resistance and echinocandin tolerance in C. parapsilosis isolates causing a large clonal candidemia outbreak among COVID-19 patients in a Brazilian ICU. Emerg. Microbes Infect. 11, 2264–2274 (2022).
Google Scholar
-
Cupozak-Pinheiro, W. J. et al. Candida species contamination in drinking groundwater from residence wells in three municipalities of midwestern Brazil and the potential human health risks. Microb. Pathog. 169, 105660 (2022).
Google Scholar
-
Prigitano, A. et al. Evolution of fungemia in an Italian region. J. Mycol. Med. 30, 100906 (2020).
Google Scholar
-
Matos, T. et al. Candidaemia in central slovenia: a 12-year retrospective survey. Mycoses 64, 753–762 (2021).
Google Scholar
-
Rodriguez, L. et al. A multi-centric study of Candida bloodstream infection in Lima-Callao, Peru: species distribution, antifungal resistance and clinical outcomes. PLoS ONE 12, e0175172 (2017).
Google Scholar
-
Xiao, M. et al. Distribution and antifungal susceptibility of Candida species causing candidemia in China: an update from the CHIF-NET study. J. Infect. Dis. 221, S139–S147 (2020).
-
Kajihara, T. et al. Distribution, trends, and antifungal susceptibility of Candida species causing candidemia in Japan, 2010–2019: a retrospective observational study based on national surveillance data. Med. Mycol. 60, myac071 (2022).
Google Scholar
-
Arastehfar, A. et al. Epidemiology of candidemia in Shiraz, southern Iran: a prospective multicenter study (2016–2018). Med. Mycol. 59, 422–430 (2021).
Google Scholar
-
Khan, Z. et al. Changing trends in epidemiology and antifungal susceptibility patterns of six bloodstream Candida species isolates over a 12-year period in Kuwait. PLoS ONE 14, e0216250 (2019).
Google Scholar
-
Beyer, R. et al. Antifungal susceptibility of yeast bloodstream isolates collected during a 10-year period in Austria. Mycoses 62, 357–367 (2019).
Google Scholar
-
Risum, M. et al. Update 2016–2018 of the nationwide Danish Fungaemia Surveillance Study: epidemiologic changes in a 15-year perspective. J. Fungi 7, 491 (2021).
-
Machado, M. et al. Incidence of candidemia is higher in COVID-19 versus non-COVID-19 patients, but not driven by intrahospital transmission. J. Fungi 8, 305 (2022).
Google Scholar
-
Routsi, C. et al. Epidemiology of candidemia and fluconazole resistance in an ICU before and during the COVID-19 pandemic era. Antibiotics 11, 771 (2022).
Google Scholar
-
Brescini, L. et al. Candidemia in internal medicine: facing the new challenge. Mycopathologia 187, 181–188 (2022).
Google Scholar
-
Barchiesi, F. et al. Factors related to outcome of bloodstream infections due to Candida parapsilosis complex. BMC Infect. Dis. 16, 387 (2016).
Google Scholar
-
Siopi, M. et al. Epidemiological trends of fungemia in Greece with a focus on candidemia during the recent financial crisis: a 10-year survey in a tertiary care academic hospital and review of literature. Antimicrob. Agents Chemother. 64, e01516-19 (2020).
Google Scholar
-
Almirante, B. et al. Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections: case–control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 44, 1681–1685 (2006).
Google Scholar
-
Escribano, P. & Guinea, J. Fluconazole-resistant Candida parapsilosis: a new emerging threat in the fungi arena. Front. Fungal Bio. 3, 1010782 (2022).
-
Cuenca-Estrella, M. et al. ESCMID guideline for the diagnosis and management of Candida diseases 2012: diagnostic procedures. Clin. Microbiol. Infect. 18, 9–18 (2012).
Google Scholar
-
Pappas, P. G. et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 62, E1–E50 (2016).
-
Rogers, T. R. et al. Molecular mechanisms of acquired antifungal drug resistance in principal fungal pathogens and EUCAST guidance for their laboratory detection and clinical implications. J. Antimicrob. Chemother. 77, 2053–2073 (2022).
Google Scholar
-
Branco, J., Miranda, I. M. & Rodrigues, A. G. Candida parapsilosis virulence and antifungal resistance mechanisms: a comprehensive review of key determinants. J. Fungi 9, 80 (2023).
Google Scholar
-
Martini, C. et al. Prevalence and clonal distribution of azole-resistant Candida parapsilosis isolates causing bloodstream infections in a large Italian hospital. Front. Cell Infect. Microbiol. 10, 232 (2020).
Google Scholar
-
Alcoceba, E. et al. Fluconazole-resistant Candida parapsilosis clonally related genotypes: first report proving the presence of endemic isolates harbouring the Y132F ERG11 gene substitution in Spain. Clin. Microbiol. Infect. 28, 1113–1119 (2022).
Google Scholar
-
Guinea, J. et al. Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med. Mycol. 60, myab068 (2021).
-
Xia, J., Huang, W., Lu, F., Li, M. & Wang, B. Comparative analysis of epidemiological and clinical characteristics between invasive Candida infection versus colonization in critically ill patients in a tertiary hospital in Anhui, China. Infect. Drug Resist. 15, 3905–3918 (2022).
Google Scholar
-
Liu, Y., Kang, M., Ye, H., Zong, Z. & Lv, X. Analysis on clinical characteristics and drug resistance of Candida parapsilosis bloodstream infections in West China Hospital, China, from 2012 to 2015. J. Mycol. Med. 28, 222–226 (2018).
Google Scholar
-
Kimura, M. et al. Micafungin breakthrough fungemia in patients with hematological disorders. Antimicrob. Agents Chemother. 62, e02183-17 (2018).
Google Scholar
-
Kimura, M. et al. Factors associated with breakthrough fungemia caused by Candida, Trichosporon, or Fusarium species in patients with hematological disorders. Antimicrob. Agents Chemother. 66, e0208121 (2022).
-
Davari, A. et al. Echinocandin resistance in Candida parapsilosis sensu stricto: role of alterations in CHS3, FKS1 and Rho gene expression. J. Glob. Antimicrob. Resist. 22, 685–688 (2020).
-
Garcia-Effron, G., Katiyar, S. K., Park, S., Edlind, T. D. & Perlin, D. S. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52, 2305–2312 (2008).
Google Scholar
-
Zhai, B. L. et al. Echinocandin heteroresistance causes prophylaxis failure and facilitates breakthrough Candida parapsilosis infection. Preprint at medRxiv https://doi.org/10.1101/2022.05.29.22275734 (2022).
-
Ricotta, E. E. et al. Invasive candidiasis species distribution and trends, United States, 2009–2017. J. Infect. Dis. 223, 1295–1302 (2021).
-
Hirayama, T. et al. Virulence assessment of six major pathogenic Candida species in the mouse model of invasive candidiasis caused by fungal translocation. Sci. Rep. 10, 3814 (2020).
Google Scholar
-
Intra, J., Sala, M. R., Brambilla, P., Carcione, D. & Leoni, V. Prevalence and species distribution of microorganisms isolated among non-pregnant women affected by vulvovaginal candidiasis: a retrospective study over a 20 year-period. J. Mycol. Med. 32, 101278 (2022).
-
Makanjuola, O., Bongomin, F. & Fayemiwo, S. A. An update on the roles of non-albicans Candida species in vulvovaginitis. J. Fungi 4, 121 (2018).
Google Scholar
-
Moreira, D. et al. Difference between the profiles presented by yeasts that colonize the vaginal mucosa or cause primary or recurrent candidiasis. Mycopathologia 186, 411–421 (2021).
Google Scholar
-
Nash, A. K. et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome 5, 153 (2017).
Google Scholar
-
Rao, C. et al. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591, 633–638 (2021).
Google Scholar
-
Martino, C. et al. Microbiota succession throughout life from the cradle to the grave. Nat. Rev. Microbiol. 20, 707–720 (2022).
Google Scholar
-
Kondori, N. et al. Candida species as commensal gut colonizers: a study of 133 longitudinally followed Swedish infants. Med. Mycol. 58, 485–492 (2020). This article reports fungal gut colonization patterns of healthy Swedish neonates during the first months of life, and how birth mode and lifestyle factors influence yeast colonization.
Google Scholar
-
Ward, T. L. et al. Development of the human mycobiome over the first month of life and across body sites. mSystems 3, e00140-17 (2018).
Google Scholar
-
James, S. A. et al. Preterm infants harbour a rapidly changing mycobiota that includes Candida pathobionts. J. Fungi 6, 273 (2020).
-
Gonia, S. et al. Candida parapsilosis protects premature intestinal epithelial cells from invasion and damage by Candida albicans. Front. Pediatr. 5, 54 (2017).
Google Scholar
-
Hamzavi, S. S. et al. Changing face of Candida colonization pattern in pediatric patients with hematological malignancy during repeated hospitalizations, results of a prospective observational study (2016–2017) in Shiraz, Iran. BMC Infect. Dis. 19, 759 (2019).
Google Scholar
-
Geros-Mesquita, A. et al. Oral Candida albicans colonization in healthy individuals: prevalence, genotypic diversity, stability along time and transmissibility. J. Oral. Microbiol. 12, 1820292 (2020).
Google Scholar
-
Benedict, K. et al. Neonatal and pediatric candidemia: results from population-based active laboratory surveillance in four US locations, 2009–2015. J. Pediatr. Infect. Dis. Soc. 7, e78–e85 (2018).
-
Lee, J. et al. Efficacy and safety of fluconazole prophylaxis in extremely low birth weight infants: multicenter pre-post cohort study. BMC Pediatr. 16, 67 (2016).
Google Scholar
-
Duzgol, M. et al. Evaluation for metastatic Candida focus and mortality at Candida-associated catheter-related bloodstream infections at the pediatric hematology–oncology patients. J. Pediatr. Hematol. Oncol. 44, E643–E648 (2022).
Google Scholar
-
Karaagac, A. T., Savluk, O. F. & Yildirim, A. I. A 5-year analysis of Candida bloodstream infections in the paediatric cardiovascular surgery ICU of a tertiary care centre. Cardiol. Young 33, 301–305 (2022).
-
Dotis, J., Prasad, P. A., Zaoutis, T. & Roilides, E. Epidemiology, risk factors and outcome of Candida parapsilosis bloodstream infection in children. Pediatr. Infect. Dis. J. 31, 557–560 (2012).
Google Scholar
-
Weimer, K. E. D., Smith, P. B., Puia-Dumitrescu, M. & Aleem, S. Invasive fungal infections in neonates: a review. Pediatr. Res. 91, 404–412 (2022).
Google Scholar
-
Cordeiro, R. A. et al. Phenotype-driven strategies for screening Candida parapsilosis complex for molecular identification. Braz. J. Microbiol. 49, 193–198 (2018).
Google Scholar
-
Zhao, Y. et al. Yeast identification by sequencing, biochemical kits, MALDI-TOF MS and rep-PCR DNA fingerprinting. Med. Mycol. 56, 816–827 (2018).
Google Scholar
-
Vecchione, A. et al. Comparative evaluation of six chromogenic media for presumptive yeast identification. J. Clin. Pathol. 70, 1074–1078 (2017).
Google Scholar
-
Sasoni, N. et al. Candida auris and some Candida parapsilosis strains exhibit similar characteristics on CHROMagar™ Candida Plus. Med. Mycol. 8, myac062 (2022).
-
Arendrup, M. C. et al. How to interpret MICs of antifungal compounds according to the revised clinical breakpoints v. 10.0 European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clin. Microbiol. Infect. 26, 1464–1472 (2020). This review discusses the importance and implications of breakpoints for antifungal resistance testing and explains the reasoning behind the revision of echinocandin breakpoints for C. parapsilosis, which now classifies wild-type isolates as susceptible instead of intermediate.
Google Scholar
-
Mikulska, M. et al. Lower sensitivity of serum (1,3)-β-d-glucan for the diagnosis of candidaemia due to Candida parapsilosis. Clin. Microbiol. Infect. 22, e645–e648 (2016).
-
Chiotos, K. et al. Comparative effectiveness of echinocandins versus fluconazole therapy for the treatment of adult candidaemia due to Candida parapsilosis: a retrospective observational cohort study of the Mycoses Study Group (MSG-12). J. Antimicrob. Chemother. 71, 3536–3539 (2016).
Google Scholar
-
Brüggemann, R. J., Jensen, G. M. & Lass-Flörl, C. Liposomal amphotericin B—the past. J. Antimicrob. Chemother. 77, ii3–ii10 (2022).
Google Scholar
-
Kontoyiannis, D. P. et al. Anidulafungin for the treatment of candidaemia caused by Candida parapsilosis: analysis of pooled data from six prospective clinical studies. Mycoses 60, 663–667 (2017).
Google Scholar
-
Qin, J. L., Yang, H., Shan, Z. M., Jiang, L. Z. & Zhang, Q. X. Clinical efficacy and safety of antifungal drugs for the treatment of Candida parapsilosis infections: a systematic review and network meta-analysis. J. Med. Microbiol. 70, 001434 (2021).
Google Scholar
-
Lass-Flörl, C. et al. Clinical usefulness of susceptibility breakpoints for yeasts in the treatment of candidemia: a noninterventional study. J. Fungi 6, 76 (2020).
-
Hoenigl, M. et al. Invasive candidiasis: investigational drugs in the clinical development pipeline and mechanisms of action. Expert Opin. Investig. Drugs 31, 795–812 (2022).
Google Scholar
-
Diaz-Garcia, J. et al. Blood and intra-abdominal Candida spp. from a multicentre study conducted in Madrid using EUCAST: emergence of fluconazole resistance in Candida parapsilosis, low echinocandin resistance and absence of Candida auris. J. Antimicrob. Chemother. 77, 3102–3109 (2022).
Google Scholar
-
Hoenigl, M. et al. The antifungal pipeline: fosmanogepix, ibrexafungerp, olorofim, opelconazole, and rezafungin. Drugs 81, 1703–1729 (2021).
Google Scholar
-
Pfaller, M. A., Carvalhaes, C., Messer, S. A., Rhomberg, P. R. & Castanheira, M. Activity of a long-acting echinocandin, rezafungin, and comparator antifungal agents tested against contemporary invasive fungal isolates (SENTRY Program, 2016 to 2018). Antimicrob. Agents Chemother. 64, e00099-20 (2020).
Google Scholar
-
Vila, T., Ishida, K., Seabra, S. H. & Rozental, S. Miltefosine inhibits Candida albicans and non-albicans Candida spp. biofilms and impairs the dispersion of infectious cells. Int. J. Antimicrob. Agents 48, 512–520 (2016).
Google Scholar
-
Bergin, S. A. et al. Systematic analysis of copy number variations in the pathogenic yeast Candida parapsilosis identifies a gene amplification in RTA3 that is associated with drug resistance. mBio 13, e0177722 (2022).
-
Yamin, D. et al. Global prevalence of antifungal-resistant Candida parapsilosis: a systematic review and meta-analysis. Trop. Med. Infect. Dis. 7, 188 (2022).
Google Scholar
-
Cornely, O. A. et al. ESCMID guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin. Microbiol. Infect. 18, 19–37 (2012).
Google Scholar
-
Marcos-Zambrano, L. J. et al. Isavuconazole is highly active in vitro against Candida species isolates but shows trailing effect. Clin. Microbiol. Infect. 26, 1589–1592 (2020).
Google Scholar
-
Arendrup, M. C., Chowdhary, A., Astvad, K. M. T. & Jorgensen, K. M. APX001A in vitro activity against contemporary blood isolates and Candida auris determined by the EUCAST reference method. Antimicrob. Agents Chemother. 62, e01225-18 (2018).
Google Scholar
-
Siopi, M. et al. Pan-echinocandin resistant C. parapsilosis harboring an F652S Fks1 alteration in a patient with prolonged echinocandin therapy. J. Fungi 8, 931 (2022).
-
Fu, L. W. et al. Different efficacies of common disinfection methods against Candida auris and other Candida species. J. Infect. Public Health 13, 730–736 (2020).
-
Kan, B. et al. Cellular metabolism constrains innate immune responses in early human ontogeny. Nat. Commun. 9, 4822 (2018).
Google Scholar
-
Hemedez, C. et al. Pathology of neonatal non-albicans candidiasis: autopsy study and literature review. Pediatr. Dev. Pathol. 22, 98–105 (2019).
-
Garcia-Gamboa, R. et al. The intestinal mycobiota and its relationship with overweight, obesity and nutritional aspects. J. Hum. Nutr. Diet. 34, 645–655 (2021).
-
Brilhante, R. S. N. et al. Exposure of Candida parapsilosis complex to agricultural azoles: an overview of the role of environmental determinants for the development of resistance. Sci. Total Environ. 650, 1231–1238 (2019).
Google Scholar
-
Lockhart, S. R. et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 64, 134–140 (2017).
Google Scholar
-
Chowdhary, A. et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 73, 891–899 (2018).
Google Scholar
-
Rybak, J. M. et al. Delineation of the direct contribution of Candida auris ERG11 mutations to clinical triazole resistance. Microbiol. Spectr. 9, e0158521 (2021).
-
Fekkar, A. et al. Hospital outbreak of fluconazole-resistant Candida parapsilosis: arguments for clonal transmission and long-term persistence. Antimicrob. Agents Chemother. 95, e02036-20 (2023). This article compares azole-susceptible clinical C. parapsilosis isolates and shows that strains harbouring the Y132F amino acid substitution are most likely to be involved in clustered and invasive infections.
-
Mesini, A. et al. Changing epidemiology of candidaemia: increase in fluconazole-resistant Candida parapsilosis. Mycoses 63, 361–368 (2020).
Google Scholar
-
Magobo, R. E., Lockhart, S. R. & Govender, N. P. Fluconazole-resistant Candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, South Africa. Mycoses 63, 471–477 (2020).
Google Scholar
-
O’Brien, C. E. et al. Identification of a novel Candida metapsilosis isolate reveals multiple hybridization events. G3 12, jkab367 (2022).
-
Arastehfar, A. et al. The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J. Fungi 6, 138 (2020). This study updates the various molecular mechanisms underlying antifungal drug resistance in Candida and Aspergillus species.
Google Scholar
-
Papp, C. et al. Triazole evolution of Candida parapsilosis results in cross-resistance to other antifungal drugs, influences stress responses, and alters virulence in an antifungal drug-dependent manner. mSphere 5, e00821-20 (2020).
Google Scholar
-
Pfaller, M. A., Carvalhaes, C. G., DeVries, S., Huband, M. D. & Castanheira, M. Elderly versus nonelderly patients with invasive fungal infections: species distribution and antifungal resistance, SENTRY antifungal surveillance program 2017–2019. Diagn. Microbiol. Infect. Dis. 102, 115627 (2022).
Google Scholar
-
Prigitano, A. et al. ICU environmental surfaces are a reservoir of fungi: species distribution in northern Italy. J. Hosp. Infect. 123, 74–79 (2022).
Google Scholar
-
Arastehfar, A. et al. Molecular identification, genotypic diversity, antifungal susceptibility, and clinical outcomes of infections caused by clinically underrated yeasts, Candida orthopsilosis, and Candida metapsilosis: an Iranian multicenter study (2014–2019). Front. Cell Infect. Microbiol. 9, 264 (2019).
Google Scholar
-
Moreno-Martinez, A. E. et al. High biofilm formation of non-smooth Candida parapsilosis correlates with increased incorporation of GPI-modified wall adhesins. Pathogens 10, 493 (2021).
Google Scholar
-
Ramos-Martinez, A. et al. Epidemiology and prognosis of candidaemia in elderly patients. Mycoses 60, 808–817 (2017).
Google Scholar
-
Arsic Arsenijevic, V. et al. Candida bloodstream infections in Serbia: first multicentre report of a national prospective observational survey in intensive care units. Mycoses 61, 70–78 (2018).
Google Scholar
-
Posteraro, B. et al. Candidaemia in haematological malignancy patients from a SEIFEM study: epidemiological patterns according to antifungal prophylaxis. Mycoses 63, 900–910 (2020).
Google Scholar
-
Klingspor, L. et al. Epidemiology of fungaemia in Sweden: a nationwide retrospective observational survey. Mycoses 61, 777–785 (2018).
Google Scholar
-
Lausch, K. R. et al. Treatment of candidemia in a nationwide setting: increased survival with primary echinocandin treatment. Infect. Drug Resist. 11, 2449–2459 (2018).
Google Scholar
-
Bretagne, S. et al. No impact of fuconazole to echinocandins replacement as first-line therapy on the epidemiology of yeast fungemia (hospital-driven active surveillance, 2004–2017, Paris, France). Front. Med. 8, 641965 (2021).
-
Adam, K. M. et al. Trends of the epidemiology of candidemia in Switzerland: a 15-year FUNGINOS survey. Open Forum Infect. Dis. 8, ofab471 (2021).
Google Scholar
-
Santolaya, M. E. et al. A prospective, multi-center study of Candida bloodstream infections in Chile. PLoS ONE 14, e0212924 (2019).
Google Scholar
-
Cortes, J. A. et al. Risk factors for mortality in Colombian patients with candidemia. J. Fungi 7, 442 (2021).
-
Rodrigues, D. K. B. et al. Antifungal susceptibility profile of Candida clinical isolates from 22 hospitals of Sao Paulo State, Brazil. Braz. J. Med. Biol. Res. 54, e10928 (2021).
Google Scholar
-
Toda, M. et al. Population-based active surveillance for culture-confirmed candidemia—four sites, United States, 2012–2016. MMWR Surveill. Summ. 68, 1–15 (2019).
Google Scholar
-
Kakeya, H. et al. National trends in the distribution of Candida species causing candidemia in Japan from 2003 to 2014. Med. Mycol. J. 59, E19–E22 (2018).
-
Xiao, M. et al. Five-year national surveillance of invasive candidiasis: species distribution and azole susceptibility from the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) Study. J. Clin. Microbiol. 56, e00577-18 (2018).
Google Scholar
-
Vasilyeva, N. V. et al. Etiology of invasive candidosis agents in Russia: a multicenter epidemiological survey. Front. Med. 12, 84–91 (2018).
Google Scholar
-
Kord, M. et al. Epidemiology of yeast species causing bloodstream infection in Tehran, Iran (2015–2017); superiority of 21-plex PCR over the Vitek 2 system for yeast identification. J. Med. Microbiol. 69, 712–720 (2020).
Google Scholar
-
Dogan, O. et al. Effect of initial antifungal therapy on mortality among patients with bloodstream infections with different Candida species and resistance to antifungal agents: a multicentre observational study by the Turkish Fungal Infections Study Group. Int. J. Antimicrob. Agents 56, 105992 (2020).
Google Scholar
-
Alobaid, K. et al. Epidemiology of candidemia in Kuwait: a nationwide, population-based study. J. Fungi 7, 673 (2021).
Google Scholar
-
Ranjbar-Mobarake, M., Nowroozi, J., Badiee, P., Mostafavi, S. N. & Mohammadi, R. Cross-sectional study of candidemia from Isfahan, Iran: etiologic agents, predisposing factors, and antifungal susceptibility testing. J. Res. Med. Sci. 26, 107 (2021).
Google Scholar
-
Reda, N. M., Hassan, R. M., Salem, S. T. & Yousef, R. H. A. Prevalence and species distribution of Candida bloodstream infection in children and adults in two teaching university hospitals in Egypt: first report of Candida kefyr. Infection 51, 389–395 (2023).
-
Kutlu, M. et al. Mortality-associated factors of candidemia: a multi-center prospective cohort in Turkey. Eur. J. Clin. Microbiol. Infect. Dis. 41, 597–607 (2022).
-
Peman, J. & Ruiz-Gaitan, A. Candidemia from urinary tract source: the challenge of candiduria. Hosp. Pract. 46, 243–245 (2018).
-
Levallois, J. et al. Ten-year experience with fungal peritonitis in peritoneal dialysis patients: antifungal susceptibility patterns in a North-American center. Int. J. Infect. Dis. 16, E41–E43 (2012).
-
Sobel, J. D. & Suprapaneni, S. Candida parapsilosis vaginal infection—a new site of azole drug resistance. Curr. Infect. Dis. Rep. 20, 43 (2018).
Google Scholar
-
Toth, R., Toth, A., Vagvolgyi, C. & Gacser, A. Candida parapsilosis secreted lipase as an important virulence factor. Curr. Protein Pept. Sci. 18, 1043–1049 (2017).
Google Scholar
-
Aboutalebian, S. et al. Molecular epidemiology of otomycosis in Isfahan revealed a large diversity in causative agents. J. Med. Microbiol. 68, 918–923 (2019).
Google Scholar
-
Fich, F., Abarzua-Araya, A., Perez, M., Nauhm, Y. & Leon, E. Candida parapsilosis and Candida guillermondii: emerging pathogens in nail candidiasis. Indian. J. Dermatol. 59, 24–29 (2014).
Google Scholar
-
Jain, A. G., Guan, J. & D’Souza, J. Candida parapsilosis: an unusual cause of infective endocarditis. Cureus 10, e3553 (2018).
Google Scholar
-
Warris, A. et al. Etiology and outcome of candidemia in neonates and children in Europe: an 11-year multinational retrospective study. Pediatr. Infect. Dis. J. 39, 114–120 (2020).
-
Trevijano-Contador, N. et al. Global emergence of resistance to fluconazole and voriconazole in Candida parapsilosis in tertiary hospitals in Spain during the COVID-19 pandemic. Open Forum Infect. Dis. 9, ofac605 (2022).
Google Scholar
-
Branco, J. et al. Clinical azole cross-resistance in Candida parapsilosis is related to a novel MRR1 gain-of-function mutation. Clin. Microbiol. Infect. 28, e1655–e1658 (2022).
-
Kim, T. Y. et al. Evolution of fluconazole resistance mechanisms and clonal types of Candida parapsilosis isolates from a tertiary care hospital in South Korea. Antimicrob. Agents Chemother. 66, e0088922 (2022).
-
Papp, C. et al. Echinocandin-induced microevolution of Candida parapsilosis influences virulence and abiotic stress tolerance. mSphere 3, e00547-18 (2018).
Google Scholar
-
Ning, Y. et al. Decreased echinocandin susceptibility in Candida parapsilosis causing candidemia and emergence of a pan-echinocandin resistant case in China. Emerg. Microbes Infect. 12, 2153086 (2022).
Google Scholar
-
Arastehfar, A. et al. Genetically related micafungin-resistant Candida parapsilosis blood isolates harbouring novel mutation R658G in hotspot 1 of Fks1p: a new challenge? J. Antimicrob. Chemother. 76, 418–422 (2021).
Google Scholar
-
Marti-Carrizosa, M., Sanchez-Reus, F., March, F., Canton, E. & Coll, P. Implication of Candida parapsilosis FKS1 and FKS2 mutations in reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 59, 3570–3573 (2015).
Google Scholar
-
Morio, F. et al. Precise genome editing using a CRISPR–Cas9 method highlights the role of CoERG11 amino acid substitutions in azole resistance in Candida orthopsilosis. J. Antimicrob. Chemother. 74, 2230–2238 (2019).
Google Scholar
-
Rizzato, C. et al. CoERG11 A395T mutation confers azole resistance in Candida orthopsilosis clinical isolates. J. Antimicrob. Chemother. 73, 1815–1822 (2018).
Google Scholar
-
Yang, F. et al. Aneuploidy underlies tolerance and cross-tolerance to drugs in Candida parapsilosis. Microbiol. Spectr. 9, e0050821 (2021).
-
Pfaller, M. A. et al. Wild-type MIC distributions and epidemiological cutoff values for amphotericin B, flucytosine, and itraconazole and Candida spp. as determined by CLSI broth microdilution. J. Clin. Microbiol. 50, 2040–2046 (2012).
Google Scholar
Acknowledgements
The authors thank S. Rofner for technical assistance in writing this article.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
M.G. has no competing interests to declare. C.L.-F. reports grant support from Gilead Sciences and Astellas Pharma; has received consulting fees or payment/honoraria for lectures, presentations and educational events from Gilead Sciences, Merck Sharp and Dohme, Pfizer, BioMerieux, F2G and Immy; and has received support for attending meetings and/or travel from Gilead Sciences.
Peer review
Peer review information
Nature Reviews Microbiology thanks Maurizio Sanguinetti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Glossary
- Antifungal susceptibility testing
-
An in vitro phenotypic method that measures the ability of a particular antifungal drug to inhibit the growth of a fungus over a range of concentrations, with the resulting minimum inhibitory concentration corresponding to the lowest concentration that inhibits fungal growth.
- Antimicrobial resistance
-
Fungal growth above established susceptibility thresholds, as determined by the minimal inhibitory concentration for a given drug.
- Clinical breakpoints
-
Values that categorize fungi as ‘susceptible’, ‘susceptible, increased exposure’ and ‘resistant’ based on minimum inhibitory concentrations, resistance markers, and pharmacokinetic and pharmacodynamic values from animal and human subjects, patient treatment and outcome data derived from controlled clinical trials.
- Clonal outbreaks
-
The occurrence of cases of diseases that are in excess of what would normally be expected and that are caused by genetically identical microbial pathogens.
- Epidemiological cut-off values
-
The highest minimum inhibitory concentrations for isolates devoid of phenotypically detectable resistance mechanisms.
- Heterozygous
-
An organism that has different copies of an allele for a particular marker.
- Homozygous
-
An organism that has the same two copies of an allele for a genomic marker.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and Permissions
About this article
Cite this article
Govrins, M., Lass-Flörl, C. Candida parapsilosis complex in the clinical setting.
Nat Rev Microbiol (2023). https://doi.org/10.1038/s41579-023-00961-8
-
Accepted: 03 August 2023
-
Published: 06 September 2023
-
DOI: https://doi.org/10.1038/s41579-023-00961-8