Blood
A new bioinformatics pipeline solves a 50-year-old blood group puzzle
Currently, a lot is known about which genes are responsible for our individual blood groups; however, not much is understood about how and why the levels of the blood group molecules differ between one person and another. This knowledge can be important for blood transfusion safety. Now a research group at Lund University in Sweden has developed a toolbox that finds the answer—and in doing so, has solved a 50-year-old mystery.
The study was published recently in Nature Communications.
For the past 30 years, the research group in Lund has studied the genetic basis of our many blood groups and their research has identified six new blood group systems. On the surface of the red blood cell are proteins and carbohydrates that are very similar between people.
However, small differences in these molecules have been shown to be due to genetic variants that encode what we know as blood group antigens. What has not been understood until now is why people with the same blood group can have different amounts of a certain blood group antigen on their red blood cells.
“This is important, because if you only have a couple of hundred blood group molecules per cell instead of a thousand or even a million molecules, then there is a risk that they maybe missed in a blood compatibility test, which can affect the safety of a blood transfusion,” explains Martin L Olsson, professor in Transfusion Medicine at Lund University, and consultant within Clinical Immunology and Transfusion Medicine, Region Skåne, who has led the project.
Since routine genetic analysis could not answer this question, the research group turned its attention to a group of proteins called transcription factors. These are molecules that can recognize different “landing” sites in DNA and work a little like a light switch to turn off/turn down genes or get them to express more strongly. Thus, transcription factors are important for the production of different proteins in the cells.
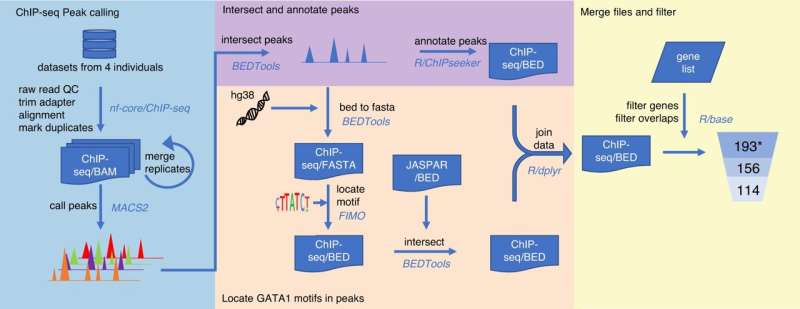
With the help of a series of bioinformatics tools (together called a pipeline) developed by Ph.D. student, Gloria Wu, the researchers could localize nearly 200 landing sites for transcription factors in 33 different blood group genes in our DNA. Then, to test the pipeline to see if the predictions were correct, the group investigated one of the most important transcription factors for red blood cell development to see if there was a genetic change in one of these landing sites. This could provide the reason why a certain blood group was downregulated to a low level.
Tested on an unsolved blood group mystery
To see how the results could be used, the researchers focused on a blood group variant called Helgeson, in which the red blood cell has unusually little of a molecule called Complement Receptor 1 (CR1), an important protein for our immune response.
The Helgeson blood group has been a mystery that has eluded the research world for a long time. Approximately 1% of the population has this blood group but it hasn’t even been possible to detect it with the help of DNA techniques. In addition, the mechanism behind the low CR1 expression remains unexplained.
“Margaret Helgeson was a medical technologist in Minneapolis in the 1970s who was trying to find compatible blood for a patient in need of a blood transfusion. Despite her best efforts she could not find any suitable blood units. In desperation, she tested her own blood, and to her surprise, found it to be a match,” recounts Jill Storry, adjunct professor of experimental transfusion medicine at Lund University and one of the researchers behind the study.
This is how the blood group became known as Helgeson. But why does a small group of people have this weak blood group? It turns out that blood donors and patients with the Helgeson blood group have a low CR1 expression because of a genetic variation in the landing site DNA sequence for an important transcription factor. This means the transcription factor cannot bind where it should and drive the production of CR1.
“Now the gene simply idles. In our study, we also showed this genetic variant to be more common in Thai blood donors compared with Swedish blood donors, which makes sense since we know from previous studies that a lower CR1 level is protective against malaria,” explains Martin L Olsson.
So, while it’s difficult to detect a lower expression of CR1 in the transfusion laboratory, it gives protection against malaria, especially in areas such as Southeast Asia where the disease is common. Thanks to this study, we now understand the mechanisms behind the Helgeson blood group and why it can be more difficult to detect in certain populations.
“Based on what we know now, we can improve the laboratory tests. Our goal is to update the existing DNA-based chip that is used for blood group tests with the new variant, which will result in a safer diagnostic test,” says Gloria Wu.
The role of blood groups in disease is the next step
With the help of this data-driven, bioinformatic pipeline, which makes it possible to get a comprehensive grip on how our blood group genes are regulated, the research group can continue to apply more of their findings to other blood groups. Furthermore, the toolbox can be utilized more widely.
“Much of our research on blood groups now uses a combination of data-based predictive tools that can point us to the right experiment to test in the lab. The next challenge is to better understand the function of blood groups by connecting the information from large databases on how diseases affect people differently depending on their blood group,” concludes Martin L Olsson.
More information:
Ping Chun Wu et al, Elucidation of the low-expressing erythroid CR1 phenotype by bioinformatic mining of the GATA1-driven blood-group regulome, Nature Communications (2023). DOI: 10.1038/s41467-023-40708-w
Lund University
Citation:
A new bioinformatics pipeline solves a 50-year-old blood group puzzle (2023, September 29)
retrieved 29 September 2023
from https://medicalxpress.com/news/2023-09-bioinformatics-pipeline-year-old-blood-group.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.