Cancer and neoplasms
Intracellular bacteria in cancer—prospects and debates
Abstract
Recent evidence suggests that some human cancers may harbor low-biomass microbial ecosystems, spanning bacteria, viruses, and fungi. Bacteria, the most-studied kingdom in this context, are suggested by these studies to localize within cancer cells, immune cells and other tumor microenvironment cell types, where they are postulated to impact multiple cancer-related functions. Herein, we provide an overview of intratumoral bacteria, while focusing on intracellular bacteria, their suggested molecular activities, communication networks, host invasion and evasion strategies, and long-term colonization capacity. We highlight how the integration of sequencing-based and spatial techniques may enable the recognition of bacterial tumor niches. We discuss pitfalls, debates and challenges in decisively proving the existence and function of intratumoral microbes, while reaching a mechanistic elucidation of their impacts on tumor behavior and treatment responses. Together, a causative understanding of possible roles played by intracellular bacteria in cancer may enable their future utilization in diagnosis, patient stratification, and treatment.
Introduction
The prospect of bacteria potentially impacting cancer gained traction in the late 19th century, with reports such as that of William Russell and colleagues suggesting that microorganisms may reside within tumors1,2. After a period of early excitement and attempts to identify a unified bacterial cause of cancer3, reproducible research on carcinogenic bacteria proved elusive for almost a century. Consequently, the bacterial theory of cancer was abandoned, and the fields of microbiology and oncology independently evolved for several decades with little intersection. In cancer research, genomic aberrations and deregulated signaling pathways have, for long, taken center stage amongst other modulating factors, as key drivers of tumorigenesis4. Conversely, vastly improved culturing and identification methods have revealed early reported “cancer microbes” to be likely contaminants5. While groundbreaking discoveries on Helicobacter pylori (H. pylori) in stomach ulcers and consequent gastric neoplasms transformed our view of microbes in cancer6, it is only in the past 15 years that the prospect of cancer microbiome research has been revisited on a larger scale7. Technological developments in next-generation sequencing (NGS) that enabled the characterization of microbiome-rich samples, such as fecal, vaginal and oral microbiomes, have revealed distinct stool microbial signatures associated with different stages of colorectal cancer (CRC)8,9,10,11 and possibly other cancers7,12.
Beyond changes in the stool microbiome, analyses of tumor sequencing data recently suggested that specific microbes may reside within tumors and their microenvironment13,14,15,16. Interestingly, some of these cancer-associated low-biomass bacteria were suggested to reside inside tumor and immune cells. Ample evidence attests to the enrichment of intracellular bacteria, such as Fusobacterium nucleatum (F. nucleatum) in CRC16,17,18. Generalization of such findings to other tumors that had been previously considered sterile is suggested by some studies14,15 and debated by others19,20, therefore meriting future confirmatory studies.
Intracellular pathogens have been extensively studied in infectious diseases. For many bacterial pathogens, intracellular localization constitutes an important, at times obligatory, component of their lifestyle. Intracellular localization bestows numerous advantages on the invading microbe, ranging from immune escape to a favorable nutritional environment and a platform for replication and dissemination. In contrast to infectious pathogens, interactions and target cells of cancer-specific intracellular bacteria remain understudied. Addressing the critical question of whether low-biomass microbial inhabitants are consistently present in tumors, and whether they bear functional implications on cancer development, progression, and therapy response is faced by multiple technical and conceptual challenges. Nevertheless, it is already yielding insightful discoveries. For example, certain bacteria detected in tumors, such as genotoxic Escherichia coli (E. coli) and enterotoxigenic Bacteroides fragilis (B. fragilis) have been linked to the production of metabolites that promote inflammation and DNA damage, contributing to the initiation and growth of tumors21,22,23. Moreover, some bacteria may influence the response of tumors to chemotherapy and immunotherapy24,25. Some of these effects may be explained by bacteria metabolizing and inactivating chemotherapeutic drugs26. Others can alter an immune response in the context of immune checkpoint blockade in modulating the response to immunotherapy27,28.
While more than a century has passed since the first reports suggesting that microorganisms may exist within cancer cells, the notion of intracellular bacteria in cancer is being revisited. As microbiology and oncology come to intersect anew, we aim to describe in this review the challenges facing the field and highlight where lessons may be learned from decades of microbiology research in a non-cancer context. Here, we provide an overview of intratumoral bacteria research, focusing on intracellular bacteria that have been suggested to colonize different tumor types and cancer models (Fig. 1). The number of studies focusing specifically on intracellular bacteria in cancer remains limited, and proper separation between intratumoral and intracellular effects is often lacking. For this reason, in this review, we refer to “intratumoral” microbes as tumor-associated microorganisms whose precise localization is unclear, while denoting them “intracellular” if they are suggested to reside within cells of the tumor microenvironment (TME). The broader relationships of the bacterial gut microbiome7,14,27,29 and other microbial kingdoms30,31 with cancer are reviewed elsewhere.
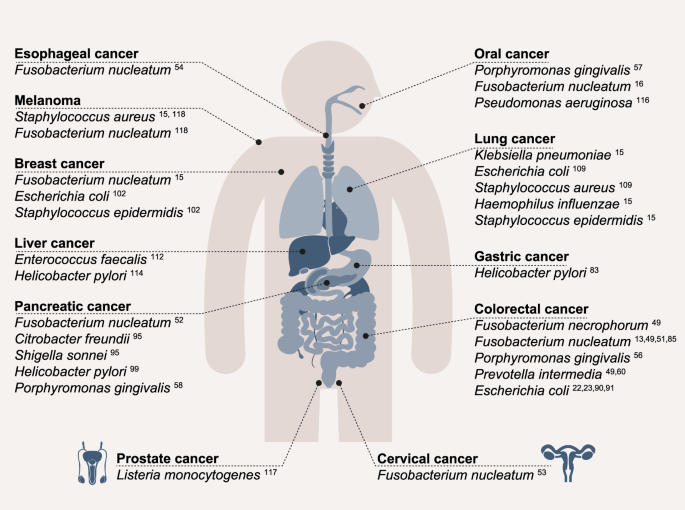
An overview of bacterial species that are suggested to be enriched in various cancers while exhibiting the capacity to invade host cells.
Lifestyles of intracellular bacteria
Intracellular pathogens have been studied for decades and yielded key insights into the mechanisms driving bacterial invasion, induction of host damage and evasion of host defense. Concepts gained from these studies, summarized below, could prove useful to the study of similar bacterial behaviors and host impacts in the cancer setting.
Strategies of bacterial invasion
Pathogen interaction with host receptors drives their adherence to host cells and triggers actin cytoskeleton rearrangements, leading to cellular invasion. Intracellular bacteria may reside in isolation, or in mixed consortia, depending on the species32,33. Bacteria that are classified as intracellular occur inside the host cell for at least parts of their lifecycle. Obligate intracellular bacteria, such as Rickettsia spp. and Chlamydia spp., have an exclusively intracellular lifestyle, while facultative intracellular bacteria, such as Salmonella spp. or Listeria monocytogenes (L. monocytogenes), can also proliferate extracellularly under certain conditions. A pathogen’s path into the host cell fundamentally varies between active invasion and passive phagocytosis, of which only active invasion will be discussed here. Bacteria can enter the host cell actively via so-called zipper or trigger mechanisms (Box 1)34. In some cases, actin polymerization and depolymerization lead to the formation of membrane protrusions, which ultimately result in the uptake of bacteria within a vacuole. Target cells of invasion are diverse and include among others epithelial cells, endothelial cells, keratinocytes, and different types of immune cells such as macrophages34,35,36,37,38,39. Intracellular localization provides bacteria with several advantages, such as protection from circulating immune cells and nutrient availability40.
Host bacterial sensing
To counter pathogenic bacterial engagement and associated cellular and tissue damage, the host has developed sophisticated systems to recognize and combat intracellular bacteria. Recognition by the innate immune system of the host is mostly mediated by pattern-recognition receptors (PRRs) such as surface-bound Toll-like receptors (TLRs) or cytosolic NOD-like receptors (NLRs) and retinoic acid-inducible gene I (RIG-I) receptors, among others. These enable constant surveillance of different compartments of the cell for signs of infection. Activation of PRRs by invading bacteria triggers a variety of inflammatory cascades, including nuclear factor kappa B (NF-κB) activation, inflammasome formation, and cytokine release, coupled with induction of an inflammation-induced infected cell death termed pyroptosis, which collectively elicit broad rejection responses to bacterial invasion41,42. In addition, upon bacterial sensing, microRNAs (miRNA) play a role in regulating various downstream processes such as cell cycle, autophagy or immune responses43 and hence can favor or combat infections. The adaptive immune system is also able to sense intracellular bacteria by host presentation of bacteria-derived peptide fragments on major histocompatibility complexes (MHC), sometimes challenged by bacterial antigenic variation44. Interaction of MHC peptide complexes with effector T cells leads to activation and proliferation of these cells, which in turn initiates differentiation and activation of other immune cells, ultimately triggering a multi-channeled antimicrobial immune response45.
Survival and proliferation strategies
To counter these defense mechanisms in a constant “arms race”, intracellular bacteria have evolved mechanisms of adaptation and resistance41. After entering the host by either the trigger or zipper mechanism, the bacteria end up in a phagosome. Phagosomes normally fuse with lysosomes to phagolysosomes, which digest their contents. Bacteria have devised intricate mechanisms of blocking this fusion, thereby escaping the phagosome, as well as other means of neutralizing bacteriolytic function, while surviving inside the host cell. For example, Mycobacterium tuberculosis (M. tuberculosis) uses an interplay of an impenetrable cell envelope, detoxification, and radicals and acquires the Ras-related protein (Rab-5A), which blocks fusion with the lysosome in enabling survival in the phagosome45. Listeria, Rickettsia, and Shigella escape from the vacuole and replicate in the cytosol46 through movement facilitated by hijacking of the host cytoskeleton via actin polymerization. Additionally, intracellular bacteria, such as Rickettsia, Burkholderia, L. monocytogenes, and Shigella flexneri (S. flexneri), can exploit the actin cytoskeleton for spreading between host cells. These pathogens utilize the host cell actin network to form protrusions into adjacent cells, leading to a double-membrane vacuole forming in the new host cell, composed of the membrane of the previous and the new host47. Other bacteria may inhibit autophagy, a degradation and recycling process that is effective against invading bacteria48. Similar invasion, evasion, and survival mechanisms may be used by intracellular bacteria in the cancer setting, and merit future studies.
Composition and localization of intratumoral bacteria
Some tumors, and in particular CRC, are convincingly shown to harbor intratumoral and even intracellular bacteria. Indeed, most CRC-associated microbes, such as F. nucleatum15,16,17,18,49,50,51,52,53,54,55, P. gingivalis56,57,58,59, and Prevotella intermedia (P. intermedia)49,60,61 are suggested to reside intracellularly. Other, traditionally considered “sterile” tumors are also suggested to feature low-biomass bacterial communities, as shown by several genomic-based14,15,20,62, and imaging-based approaches15,16. Of note, some of the genomic approaches utilized by these studies have been recently challenged19,63, with a definite resolution of these debates, and high-resolution characterization of intratumoral bacteria meriting future studies.
NGS, ranging from characterization of the 16S rDNA gene to global characterization of the bacterial pan-genome using shotgun metagenomic sequencing, is extensively utilized in unraveling fecal cancer-associated microbiome signatures at low host DNA contamination levels8,9. In contrast, efforts to profile the low-biomass tumor-resident microbiome prove to be more challenging, given a substantial excess of human reads, imperfections in reference databases, experimental and computational contaminations, and severe batch effects caused by varying collection methods, sample processing, and sequencing pipelines14,62,64. Suggested solutions include novel low-biomass sequencing approaches, in which five regions of the 16S rRNA gene are simultaneously amplified and sequenced, enabling higher coverage and resolution15,65. These amplicons can then be computationally combined using Short MUltiple Regions Framework (SMURF)65, a platform built specifically for this method. Of note, 5 region 16S rRNA sequencing is highly sensitive and bacterial DNA can be found in many sources of contamination, making it challenging to distinguish between low-biomass bacterial signals originating from the tissue and those originating from environmental contamination66,67. Such distinction requires careful assessment by multiple controls to bioinformatically filter out interfering signals15. Additional efforts to improve disentangling of true low-biomass signals from contamination and noise focus on improved harmonization of experimental and computational pipelines68, the use of multiple technical and biological controls, and validation of sequencing-based results with additional, non-genomic modalities. With these modalities providing some solution to the low-biomass microbial challenges, future development is needed to further disentangle true microbial signals from noise.
Metatranscriptomic analysis can provide further functional insights into microbial consortia and their relationships with their host69. Dual RNA sequencing approaches have emerged in 201232 to gain insights into simultaneous gene expression of host cells, invading bacteria and their interactions32. These modalities have evolved to enable the assessment of co-infections of different bacterial species, and even viruses and bacteria, in decoding inter-species and inter-kingdom interactions70. Single-cell RNA sequencing may further minimize bulk contamination by cellular and genetic heterogeneity and reveal the extent to which individual cells are targeted and shaped by invading bacteria. For example, Galeano Niño et al. established a method called invasion–adhesion-directed expression sequencing (INVADEseq), focusing on cell attachment and invasion with spatial resolution. In INVADEseq, primers targeting a conserved region of the bacterial 16S rRNA locus enable the generation of cDNA libraries containing bacterial transcripts from human cells associated with bacteria. While good resolution may be achieved, distinction between invaded and cell-adjacent bacteria still remains a major obstacle16. Using this method in oral squamous cell carcinoma (OSCC), Fusobacterium and Treponema colonization were associated with macrophages and aneuploid epithelial cells16,71. Another tool, Single-cell Analysis of Host–Microbiome Interactions (SAHMI)52, relies on recovering and denoising microbial signals from single-cell sequencing data of the host tissue and mapping it to a microbial reference genome. In addition, computational prediction tools such as Host–Microbe Interaction PREDictor (HMI-PRED), used for prediction of protein–protein interactions, may enable further elucidation of host–microbe interactions72.
Imaging techniques are increasingly used to complement genomic low-biomass microbial signals15. These include immunohistochemistry (IHC), FISH and high-resolution electron microscopy. The recently introduced correlative focused ion beam/scanning electron microscopy (c-FIB/SEM) combines volume electron microscopy and fluorescence microscopy, thereby enabling better understanding of host-microbe interactions at the 3D ultrastructural level73. Utilization of fluorochrome-conjugated bacteria-specific antibodies coupled with permeabilization of host samples may enable intracellular microbial imaging18,74. However, killing of both bacteria and host cells limits the use of this approach to endpoint assays. Stable labeling of intracellular bacteria, such as F. nucleatum, is challenged by difficulties in genetic modification, despite recent progress75. Bacterial chemical labeling50,76, while beneficial for short-term analyses, suffers from attenuation of the signal by serial dilution, promoted by cell division over time. Fluorescently labeled d-alanine may offer a solution to this challenge, as it is metabolically incorporated into the bacterial cell wall and thus may enable the detection of living, metabolically active tumor-associated bacteria upon incubation with fresh tumor samples77.
Recent work from the Bullman group incorporated several state-of-the-art spatial profiling methods in better characterizing bacterial niches in tumors16. RNAscope-fluorescence in situ hybridization (RNAscope-FISH) enabled the visualization of bacterial RNA within individual host cells, thereby revealing locations of bacteria within tumors. Using the digital spatial profiling platform GeoMX, expression of 77 proteins related to anti-tumor immunity was correlated with tumor characteristics. Through application of 10X Visium spatial transcriptomics, positional information could also be documented in tumor niches. Using these methods, it was suggested that most intratumoral bacteria are located in micro-niches at the tumor margin, featuring an immunosuppressive and poorly vascularized microenvironment16.
Suggested intracellular bacteria across tumor and tissue types
Studies utilizing the above technologies suggest that tumors may harbor unique microbial signatures. The most extensive evidence demonstrating such tumor-residing bacteria and their tumor-modulatory function involves gastric and colorectal cancer. A definite level of proof regarding presence, extent, and possible functions of intratumoral microbiomes, and the prospect of their utilization in cancer diagnosis14, remains debated19 and awaits future validation.
Gastric cancer
H. pylori was the first, and currently only bacterium recognized by the World Health Organization as a carcinogen to date. H. pylori constitutes a prototypical driver of gastritis and subsequently gastric cancer via diverse mechanisms such as induction of chronic inflammation78,79,80 and modulation of wingless-related integration site (Wnt) signaling81, amongst others. The many direct and indirect contributions of H. pylori to gastric cancer development have been reviewed elsewhere82. Of interest to intratumoral microbiome research, some of the effects conferred by H. pylori have been suggested to be linked to attachment and potential invasion of gastric epithelial cells74,75, supported by the presence of H. pylori in gastric cancer tissue83.
Colorectal cancer
Colorectal cancer is the third most common cancer worldwide and the second leading cause of cancer deaths in the US84. Due to its proximity to a rich luminal microbiome, it is unsurprising that it has been studied extensively for intratumoral and intracellular bacteria. As noted above, among the CRC-associated bacteria with intracellular localization, F. nucleatum is the best studied species to date. Several studies have detected an enrichment of F. nucleatum, in human adenomas and CRC compared to healthy colon tissue samples13,51,85. The invasive behavior of patient-derived F. nucleatum subspecies was validated by functional assays in CRC cell lines49,74,86. Intracellular F. nucleatum is also enriched in inflammatory bowel disease (IBD), a condition predisposing to CRC, especially in patients suffering from an active disease compared to those in remission87. F. nucleatum strains from patients featured an invasive behavior in 2D cell line assays, which correlated with IBD status74. Fusobacterium necrophorum (F. necrophorum), another member of the Fusobacterium genus with an invasive behavior88, has also been associated with CRC49. Another periodontal-linked bacterium, P. gingivalis, was enriched in CRC compared to healthy adjacent tissue56 and suggested to reside intracellularly16 while featuring an invasive behavior89. P. gingivalis dominantly occurs in the oral cavity, where it resides in gingival epithelial cells and is linked to periodontitis and OSCC57,89. Comparing paired adenocarcinoma and polyp samples, a higher abundance of P. intermedia was detected in adenocarcinoma samples60, and it was suggested to reside, at least partly, intracellularly16,49,61. Pathogenic strains of E. coli are also recurrently found in CRC21,22,23,90. Some of those are able to thrive within macrophages91 and could be cultured from tumor tissue after gentamycin treatment92, suggesting that they may feature a capacity to intracellularly survive. Of note, a recent report suggests a close interplay between attachment and genotoxicity for some of these strains92.
Pancreatic cancer
Pancreatic ductal adenocarcinoma (PDAC) constitutes the most common form of pancreatic cancer, accounting for more than 85% of all cases93. In PDAC, Campylobacter, Leptotrichia, F. nucleatum and Clostridioides difficile were suggested as intratumoral species based on single-cell sequencing data52. Further studies detected Citrobacter freundii and Shigella sonnei94 as part of the pancreatic cancer TME, while other studies suggested that these bacteria occur intracellularly95,96. A meta-analysis from 2011 concluded that H. pylori, which was shown to be capable of invading host cells97, enhances the risk of pancreatic cancer98. P. gingivalis has also been reported to be dominantly enriched in patients with pancreatic adenomas and possibly correlated with PDAC disease progression58. PDAC was also suggested to bear fungi15,99,100, but some of these findings were recently contested63, meriting future research.
Other cancer types
F. nucleatum15, Staphylococcus epidermidis (S. epidermidis), E. coli101, and other members of the genus Staphylococcus, Streptococcus, and Lactobacillus102 were suggested to be present in human breast cancer samples. The abundance and potential intracellular localization of microbes in breast cancer awaits definite resolution. F. nucleatum was suggested to be enriched in cervical cancer53 and esophageal cancer54 compared to adjacent healthy tissue. Associations and potential functional roles for intracellular Chlamydia spp. have been described in cervical and ovarian cancer103,104,105. This pathogen has been shown to decrease p53 signaling and DNA damage response106, in facilitating the pathogen’s intracellular replication, induction of ROS production and DNA damage107. Analysis of a TCGA lung adenocarcinoma dataset, a subtype of lung cancer, suggested that E. coli and Staphylococcus aureus (S. aureus) may reside within these tumors108. S. epidermidis, Haemophilus influenzae (H. influenza), and Klebsiella pneumoniae (K. pneumoniae)15, were suggested to reside in lung cancer samples in separate studies109. Using transmission electron microscopy, intracellular bacteria could be visualized in intrahepatic cholangiocarcinoma, a form of liver cancer, and surrounding tissue, but no further characterization of the bacteria was performed110. A presence of Enterococcus faecalis (E. faecalis) was demonstrated in hepatocellular carcinoma samples and suggested to have a prominent role in liver carcinogenesis111, also in the in vivo setting112. In hepatocellular carcinoma, the presence of H. pylori was detected only in cancer samples, while it was absent in healthy controls113. Based on spatial analysis, Galeano Niño et al. revealed that the genera Parvimonas, Peptoniphilus and Fusobacterium were most abundant in an oral cancer subtype, OSCC16. Pseudomonas aeruginosa, which is capable of an intracellular lifestyle114, was detected in the same cancer subtype115. L. monocytogenes was detected in the prostate cancer TME116, while S. aureus was suggested to be present in melanoma15,117. It is noteworthy that in these studies, low-biomass approaches utilized different protocols, isolation methods and sample handling techniques, which can collectively lead to variations and inconsistencies between studies. For example, B. fragilis was suggested to be more abundant in CRC in one study118, while other studies reported no significant differences in its abundance119, which could stem from differences in participant characteristics and varying experimental designs. Given recent debates, these low-biomass findings merit future validation.
Synergies between intracellular bacteria
Many microbiome-modulated diseases are not driven by single pathogens, but rather by synergistic consortia that can evolve into a dysbiotic state that impairs host homeostasis via concerted activities120. For example, gingival P. gingivalis infection in germ-free mice induces periodontal disease only in the presence of commensal communities, through joint promotion of polymicrobial biofilms via regulation of cytokine levels121. Similar concepts could also be relevant in the cancer context. Initial studies show that F. nucleatum-positive CRC tissues featured non-random co-colonization with commensals such as B. fragilis or P. intermedia, whereas tissues lacking F. nucleatum featured different bacterial colonization patterns49. These co-colonization patterns may bear functional importance. For example, communities containing both F. nucleatum and P. gingivalis featured a higher rate of invasion of gingival epithelial cells as compared to those featuring either of these species without the other122. Further investigation is needed to determine the extent to which bacterial composition and co-occurrence patterns causally shape their modulatory activities impacting cancer.
Routes of bacterial colonization into tumors
Routes by which intratumoral bacteria may reach and persist in the TME remain elusive to date. It is conceivable that bacteria from the adjacent normal tissue become enriched at tumor sites during tumorigenesis due to the changed microenvironment and easier tissue access upon disruption of epithelial and mucus barriers, for example, the gastrointestinal tract. For many other tumor types and even most gastrointestinal tumors, additional routes are plausible, including transmission of bacteria to distant sites via the bloodstream or the lymphatic system123,124.
Oral cavity-to-gut translocation of bacteria constitutes an active field of investigation in CRC and other cancer research. Bacteria of the oral cavity are involved in local inflammatory diseases such as periodontal disease and are also enriched in various, seemingly unrelated, tumor tissues. Intriguingly, a 10-year study found periodontal patients to have an increased risk of cancer development, particularly pancreatic cancer125, but also other cancers such as lung cancer, head and neck cancer, abdominal and esophageal cancer, breast cancer, and CRC126. Several periodontal bacteria, such as F. nucleatum and P. gingivalis, feature an invasion and colonization capacity even in their gingival tissue of origin127,128. Elucidation of effector similarities of such oral microbiome bacteria upon migration into tumor sites merits future mechanistic research. F. nucleatum, a well-studied invasive cancer-related bacterium, is believed to migrate from its natural habitat within the oral cavity124 via the bloodstream. From the bloodstream, local enrichment in colorectal adenocarcinoma is suggested to be mediated by binding between bacterial fibroblast activation protein-2 (Fap2) and host epithelial d-galactose-β(1-3)-N-acetyl-d-galactosamine (Gal-GalNAc), which is overrepresented at CRC sites129. In addition, F. nucleatum can bind to the salivary protein statherin via FomA, which functions in biofilm formation and is known to bind especially to P. gingivalis51,130. In PDAC, gut bacteria can migrate from the anatomically connected upper gastrointestinal tract into the tumor131. Of note, translocation of gut microbes to sites like the liver can precede tumor formation111. After entering the TME, factors such as nutrient-rich niches, low pH, necrotic foci, hypoxia, or abundant blood supply may support bacterial colonization132,133,134. Similar considerations may apply to microbial colonization of metastases. In a murine breast cancer model, bacteria detected in circulating tumor cells were also enriched in lung metastasis sites, suggesting that some bacteria, and specifically intracellular bacteria, may migrate to metastatic sites within tumor cells via the systemic circulation102. Further evidence suggests that certain strains of E. coli can disrupt the gut vascular barrier and support CRC cells at multiple stages of their metastatic dissemination135.
Potential impacts of intracellular bacteria on cancer phenotypes
The above observations, suggestive of the occurrence of tumor-associated bacteria, constitute an intriguing starting point of investigation of potential causation, e.g., whether tumor-residing or even intracellular bacteria may impact tumorigenesis, cancer progression, and treatment responsiveness. This “chicken-and-egg” dilemma constitutes a formidable challenge that requires sophisticated experimental models and techniques to detect causes and mechanisms.
Bacterial invasion models
To mechanistically study bacterial invasion, in vitro models of varying complexity are often combined with antibiotic protection assays that use non-penetrant antibiotics, such as gentamicin, to enrich viable intracellular microbes86,136,137,138,139,140,141. There are different approaches and models to study invasion that vary in their ease of use and the representation of in vivo physiology, a summary of which is depicted in Fig. 2. One of the most commonly used invasion assays is based on the co-incubation of 2D cell lines, such as Caco-2 cells derived from CRC, with microbes that actively invade the host cells18,25,50,74,86,138,139. Such 2D studies revealed important interactions between surface components of bacteria and the host, invasion, and the induction of oncogenic and inflammatory pathways upon infection18,50,86,129. For example, the use of such platforms uncovered the importance of the lectin Fap2, expressed on F. nucleatum, binding to Gal-GalNAc on host cells. This interaction, in turn, drives an accumulation of Fusobacteria in CRC50,129, upregulation of inflammatory markers upon invasion13,18,50, and direct modulation of the TME138. Moreover, F. nucleatum is suggested to enhance CRC progression through its FadA adhesin, which attaches to epithelial cadherin (E-cadherin) and may activate the Wnt/β-catenin signaling pathway86 leading to enhanced proliferation86. Such 2D invasion assays have also proved invaluable in testing means to interfere with bacterial tumor cell invasion. For example, galactosides were discovered to interfere with Fap2-mediated invasion of F. nucleatum into CRC cells50. However, many proposed cancer-associated bacteria thrive in anaerobic conditions, making it difficult to mimic a suitable 2D environment that would phenocopy their in vivo niche. In addition, 2D in vitro invasion models remain limited to timeframes of a few hours, which may be too short for the effects of viable intracellular bacterial invasion and related metabolic crosstalk to become apparent142,143,144, while the immortalized character of most cell lines makes it difficult to elucidate differences between healthy and cancer cells.

Key features of cell-based models with varying complexity focusing on the study of bacterial invasion, the physiological environment, and model characteristics. Advantages and disadvantages for multiple parameters are compared and evaluated by a traffic light system.
To overcome these limitations, new models enable more physiologically relevant conditions for such invasion studies, which are summarized in Fig. 2. Organoids represent a useful model to study pathogen-host interactions because they mimic the polarized epithelial cell layer enclosing a luminal compartment138,139. There are various ways to use organoids as co-culture platforms, including microinjection of bacteria into the hypoxic lumen from where they can actively invade the surrounding epithelial layer in the right orientation. Early steps of invasion of bacteria such as Salmonella enterica serovar Typhimurium (S. Typhimurium) were decoded using such platforms. Further usage of invasion platforms, including spheroids145, and transwell invasion assays146 is likely to reveal additional functional impacts of intracellular bacteria in cancer.
Organs-On-Chip (OOC) models, like CRC OOCs, enable the study of bacterial tumor invasion at an enhanced level of complexity, by replicating environmental features, including stretch, flow, and even complex gut microbial compositions76,147,148. Using Caco-2 colonized OOCs149, factors such as crypt-like structures, peristalsis, and flow were shown to have a substantial impact on Shigella invasion. Mouse models can further elucidate the impacts of intracellular and intratumoral bacteria on tumor growth in a complete host environment. As one example, F. nucleatum-positive colorectal tumors were subcutaneously transplanted into immunodeficient mice and tracked over time. Using this model, viable F. nucleatum could be maintained over time, while antibiotic administration reduced tumor growth49. In the C57BL/6 Apcmin/+ CRC mouse model, increased tumor formation occurred upon gavage of F. nucleatum, while demonstrating a key role of FadA in its tumor-promoting activity140. Enhanced tumorigenesis could be likewise demonstrated upon transplantation of the P. gingivalis pre-infected pancreatic cancer cell line PANC1, suggested to be mediated by enhancement of phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling128.
Modulation of immune responses
Infection of host cells often generates potent immune responses. For example, endothelial cells invaded by the obligate intracellular bacterium Orientia tsutsugamushi elicit an interferon response69 promoting immune cell recruitment150. S. Typhimurium that infects macrophages can replicate intracellularly, resist host reactive oxygen species (ROS) responses and actively induce macrophage polarization150. Within the cytosol, F. nucleatum can be sensed via PRRs such as RIG-I151 or alpha kinase 1 (ALPK1), leading to NF-κB activation. This leads to various signaling cascades such as expression of the proinflammatory cytokines interleukin (IL)-6, IL-8, and tumor necrosis factor alpha (TNF-α), upregulation of adhesion molecules152, enhanced proliferation or autophagy51,151. Suggested mechanisms of intra- and extracellular F. nucleatum affecting proliferation, autophagy and TME modulation of the host are depicted in Fig. 3. Autophagy is modulated by various intracellular bacteria, affecting their niche and enabling intracellular survival. F. nucleatum, for example, may promote initiation of autophagy through downregulation of miR-18a* and miR-480243, while autophagosome fusion with the lysosome may be inhibited via intracellular F. nucleatum-mediated upregulation of miR-3148,153. An expansion of myeloid-derived immune cells including tumor-associated macrophages (TAMs), tumor-associated neutrophils (TANs) and myeloid-derived suppressor cells (MDSCs) in a F. nucleatum-inoculated CRC mouse model was accompanied by T-cell suppression and increased expression of immunosuppressive molecules such as CTLA4 and arginase-117,154. Recruitment of myeloid cells by intratumoral bacteria may also contribute to inflammation via activation of the Janus kinase/signal transducer and activator of transcription (JAK-STAT) pathway and secretion of interleukins and chemokines87. Modulation of natural killer (NK) cells by bacteria in the TME can further promote an immunosuppressive environment. For example, interaction of Fap2, expressed by F. nucleatum, with the human inhibitory receptor T-cell immunoreceptor with Ig and ITIM domains (TIGIT) expressed on immune cells such as NK cells and T cells, abrogated NK cell-mediated killing of human tumor cells138. Similarly, F. nucleatum-induced Fap2-dependent mechanisms can drive lymphocyte apoptosis155,156. Tumor regions featuring a high bacterial load may correlate with reduced vascularization, thereby contributing to the formation of an immunosuppressive environment16. Intratumoral bacteria may also modulate the TME adaptive immune response via the presentation of bacteria-derived peptide fragments on host antigen-presenting cell HLA complexes. Indeed, in melanoma, peptides of species such as F. nucleatum and S. aureus were presented on TME antigen-presenting cells. Some of these peptides appear to be potentially immunogenic, thereby bearing a capacity to activate adaptive immune responses while promoting tumor infiltration of lymphocytes and generating an inflammatory immune response117. Similar findings are emerging in glioblastoma multiforme157, where multiple bacteria-derived antigens were suggested to be recognized by T cells of the TME. However, several of the suggested microbe-harboring tumors appear to be cold, or relatively immunotolerant. This apparent lack of tumor antibacterial immune reactivity, despite the suggested presence of highly immunogenic bacteria, merits future studies.

The opportunistic pathogen F. nucleatum impacts host cell behavior via numerous extra- and intracellular mechanisms. F. nucleatum interacts with the host via protein interactions such as Fap2-Gal-GalNAc or FadA-E-cadherin and can invade cell types such as epithelial cells, endothelial cells or macrophages. Intracellular F. nucleatum is sensed by membrane-bound and cytosolic PRRs, mediating NF-κB activation and expression of inflammatory molecules such as precursors of IL-1b and IL-18 that are activated by cleavage by inflammasomes and then released from the host cell. Activation of cytosolic PRRs also affects increased expression of ICAM-1. The binding of LPS to TLR4 causes upregulation of autophagy, linked to chemoresistance. F. nucleatum also leads to the upregulation of miR-31 which inhibits the fusion of autophagosomes with the lysosome and enables persistent infection. Consequences of F. nucleatum invasion include increased JAK-STAT signaling, enhanced secretion of proinflammatory markers, EMT phenotype and bacterial antigen presentation on HLA molecules. The binding of FadA to E-cadherin influences downstream Wnt signaling, leading to enhanced proliferation. Extracellularly, F. nucleatum induces secretion of myeloid chemoattractants that recruit TANs, DCs and TAMs, leading to suppressed activity of CD4+ T cells. Direct interaction of F. nucleatum with immune cells via Fap2-TIGIT binding, expressed on T and NK cells, mediates decreased cytotoxicity and ultimately inhibited killing activity. CCL20 C-C Motif Chemokine Ligand 20, CTLA4 cytotoxic T-lymphocyte associated protein 4, CXCL C-X-C motif chemokine ligand, DCs dendritic cells, E-cadherin epithelial cadherin, Fap2 fibroblast activation protein-2, F. nucleatum Fusobacterium nucleatum, Gal-GalNAc d-galactose-β(1-3)-N-acetyl-d-galactosamine, IFN interferon, ICAM-1 intercellular adhesion molecule 1, IL interleukin, JAK Janus kinase, LPS lipopolysaccharide, miRNA-31 microRNA-31, NF-κB nuclear factor kappa-light-chain-enhancer of activated B cells, NK natural killer, NLRP3 NLR family pyrin domain containing 3, PRRs pattern-recognition receptors, STAT signal transducer and activator of transcription, TAMs tumor-associated macrophages, TANs tumor-associated neutrophils, TIGIT T-cell immunoreceptor with Ig and ITIM domains, TLRs Toll-like receptors, TNF tumor necrosis factor, Wnt wingless-related integration site.
Impact on tumor metastasis
Intratumoral and intracellular microbes may also regulate the metastatic cascade. Whole genome sequencing revealed a similarity of 99.9% between F. nucleatum isolates from primary CRC and liver metastasis, indicating likely bacterial migratory characteristics between these tumor sites49. Microbial regulation of the metastatic process may involve a variety of mechanisms. Epithelial–mesenchymal transition (EMT) is a dynamic process of epithelial conversion to a mesenchymal phenotype, characterized by a gradual loss of epithelial features such as epithelial cell adhesion molecule (EpCAM) expression or strong cell-cell contacts158. F. nucleatum may induce transcriptional features of EMT such as upregulation of Vimentin, Snail or Slug16,159,160. Higher intratumoral F. nucleatum abundance is associated with an increase in ALPK1, which in turn results in upregulation of intercellular adhesion molecule 1 (ICAM-1) on the host cell surface. This leads to increased attachment of CRC cells to endothelial cells which is suggested to promote EMT and ultimately metastasis152. Other suggested microbial mechanisms impacting host tumor cell metastatic potential include the secretion of bioactive products, as exemplified by F. nucleatum-derived short-chain fatty acid formate that was recently linked to CRC cell stemness and invasion161. In addition, F. nucleatum may induce the expression of pro-migratory and pro-metastatic genes in a CRC cell line, impact the motility of cancer cells, and modulate the virulence factor Fap2-driven host cell binding and suppress T-cell infiltration16. Keratin7 (KRT7) is reported to be upregulated in tumors upon F. nucleatum presence and was suggested to enhance lung metastasis in a murine CRC model162. In addition, tumor-associated bacteria may modulate cell-cycle signaling pathways16 and induce senescence in the host. Unlike apoptotic cells, senescent cells, which undergo irreversible cell-cycle arrest, remain viable. The senescence-associated secretory phenotype (SASP) involves the release of factors, which promote inflammation and malignancy (Fig. 4). In CRC, invasive P. gingivalis is enriched and correlated with higher levels of butyrate, possibly inducing SASP59. Regarding the secretion profile, F. nucleatum invasion also induced increased exosome secretion of CRC cells carrying metastasis-related miRNAs and proteins, which, in turn, were internalized by uninfected cells and activated Wnt/β-catenin signaling, enhancing migration and proliferation163.

Bacteria colonizing host tumor cells may alter a multitude of cancer-related features. These include gene expression changes triggered by host bacterial sensing, SASP or alterations in proliferation, and induction of metastasis-linked behavior such as migration, invasion, EMT or release of exosomes. Intracellular bacteria may modulate autophagy, immune cell recognition and antigen presentation, potentially impacting cancer immune recognition. Ultimately, bacteria-driven host modulations can lead to alterations in drug metabolism and consequently therapy response, thereby impacting the outcome of cancer therapies. EMT epithelial–mesenchymal transition, SASP senescence-associated secretory phenotype.
Effects on drug metabolism and treatment responsiveness
In addition to their ability to disrupt endogenous cellular processes, bacteria can metabolize drugs, thereby potentially affecting tumor responses. Several studies suggest that bacteria-mediated drug metabolism may induce drug activation or promote adverse effects. For example, gut bacteria-derived beta-glucuronidase can metabolize the chemotherapy irinotecan in a manner promoting increased mucosal toxicity164. In a larger in vitro screen in which 30 FDA-approved drugs were co-incubated with bacteria, 10 compounds were found to feature a decreased efficacy, while 6 featured an increased efficacy165. Geller et al. reported that Gammaproteobacteria may confer gemcitabine resistance to CRC cell lines by metabolizing the drug to its inactive form by the bacterial enzyme cytidine deaminase. Consequently, antibiotic treatment promoted a regained responsiveness to gemcitabine in a CRC mouse model. Importantly in this study, the most common species in human PDAC belonged to the Gammaproteobacteria class, while 14 out of 15 PDAC-derived bacterial cultures mediated gemcitabine resistance upon co-culture with CRC cell lines. Recently, E. coli-mediated inactivation of the nucleoside analog 5-fluorouracil (5-FU) was associated with a poorer therapy response. Interestingly, 5-FU was also suggested to inhibit the growth of F. nucleatum166, which might further contribute to its therapeutic activity. Neoadjuvant chemoradiotherapy with fluoropyrimidines (which are activated to 5-FU) decreased the prevalence of F. nucleatum from 58 to 26% in locally advanced rectal cancers, while the persistence of F. nucleatum following treatment predicted a relapse167. Co-cultivation of 5-FU and Oxaliplatin with F. nucleatum suggested that an altered chemotherapeutic drug response was linked to a decrease in two miRNAs (miR-4802 and miR-18a*) driving enhanced expression of autophagy-related proteins25. Understanding the effects of microbial colonization in tumors on treatment effectiveness and adverse effects may enable tailoring treatment to the individual based on their microbial profiles, in optimizing dosing while minimizing adverse effects.
Targeting intracellular bacteria in cancer treatment
Intratumoral bacteria may represent potential therapeutic targets in cancer treatment. For example, antibiotic treatment of mice transplanted with F. nucleatum-positive patient-derived CRC xenografts reduces tumor size and cancer cell proliferation, indicating that bacterial suppression may support tumor growth suppression49. However, targeting intratumoral pathogens remains challenging, as in many cases internalized antibiotics do not reach the minimum inhibitory concentration in the TME168, while high doses are associated with adverse effects on the host and its microbiome. Currently tested intracellularly acting antimicrobials include sulfonamides, quinolones, tetracyclines, and beta-lactams169. However, emerging antibiotic resistance requires the development of further therapeutic strategies that overcome low cellular permeability and microbial resistance. Explored modalities include intracellularly acting antimicrobial peptides (AMPs), such as fungal plectasin, which suppresses intracellular methicillin-resistant S. aureus (MRSA). Nonetheless, AMP activity may be abrogated upon reaching the host cell cytoplasm. Cell-penetrating peptides (CPP) can translocate to the nucleus, mostly via endocytic pathways, where they may exert antibacterial effects. CPP can also be used as translocation backbones to direct drugs into infected cells. As an example, the CPP Tat was coupled to the antibiotic gentamicin, leading to a reduction of intracellular E. coli K1170. Singh et al. proposed to conjugate the antibiotics polymyxin B sulfate and sushi peptide to gold nanoparticles, in effectively targeting intracellular Salmonella typhi171. Utilization of bacteriophages represents another experimental approach in specifically eliminating tumor-associated bacteria. For example, phages targeting F. nucleatum, combined with irinotecan were able to reduce tumor growth, while selectively inhibiting CRC-associated bacterium F. nucleatum in vivo172. Similarly, phages directed against adherent invasive E. coli reduced tumor burden in a CRC mouse model173. These pioneering efforts may be further optimized by the use of phage combinations suppressive target bacterial strains through different receptors, as was recently shown in the context of IBD174. However, phage treatment of intracellular bacteria is challenged by limited access into host cells, a changed environment altering intracellular phage-bacterial engagement, and the necessity for phages to transit between host cells in targeting multiple intracellular bacteria.
Utilization of intracellular bacteria in cancer treatment
Intracellular bacteria may be potentially utilized as medical interventions or treatment delivery vectors. For example, Salmonella used as a nonpathogenic delivery system targeting therapeutic proteins into the cytoplasm induced a reduced tumor growth and metastasis in mouse models175. Modification of the Photorhabdus asymbiotica type VI secretion system, which plays a role in macrophage invasion and intracellular survival, enabled the delivery of custom payloads into human cells176,177,178. Additionally, intratumoral bacterial peptide human leukocyte antigens (HLA) presentation on host cells may enable the harnessing of antibacterial immunity in cancer immunotherapy. As the bacterial antigens are recognized as non-self, they could serve as targets for immunotherapy stimulating an immune response117. In 2004, an attenuated S. Typhimurium systemically administered to a mouse model of melanoma featured a sustained ability to invade tumor cells. Prior vaccination of mice that generated Salmonella-specific T cells resulted in a substantial improvement in tumor load. Tumor cell invasion was suggested to play a role in this effect, as the use of non-invasive S. Typhimurium resulted in a marked attenuation of the anti-tumor response179.
Limitations and challenges in the study of intracellular tumor-residing bacteria
Despite recent advances in our collective understanding of the possible presence and activities of microbes in cancer, the field remains in its infancy and faces major challenges and obstacles. While the intracellular presence of Fusobacterium is well documented15,16,17,18,49,50,51,52,53,54,55, the presence of microbial communities, consisting of bacteria14,15,62 and fungi14,15,99 within different tumors that were commonly considered sterile has been recently debated19,63,180 and merits definite characterization by future studies. Such low-biomass tumor microbiome characterization is challenged by a variety of technical and biological challenges and potential biases stemming from contaminations, batch effects, erroneous read allocation, and imperfection in analytical pipelines, while lacking resolution to identify intracellular localization of bacteria in conjunction with tumor phenotype measurements. Examples of the challenges facing such research are presented in Box 2. The study of bacterial invasion using animal models and organoids will further contribute to our understanding of the true extent of intratumoral bacterial colonization and its potential functions. In such future experiments, the importance of addressing experimental and computational contaminations through the use of robust and diverse controls and analytical tools cannot be overstated and is highlighted by the long-standing debate as to the existence of a placental microbiome which has only recently been resolved (as likely not being present) by a large consortium effort181.
Disentangling the effects of extracellular microbes from those of true tumor cell-penetrating intracellular bacteria may enable to quantify their true impact. Importantly, recent advances have highlighted the likely heterogeneous distribution of bacteria between different specimens and even within the same sample16. Despite advances in bacterial identification, variations on a species and subspecies level are often not easy to decode, especially in the low-biomass microbial setting, with different strains often grouped solely by their genus, disregarding their diversity and presence of virulence factors in specific strains.
An additional major challenge stems from the crucial need to transform our level of understanding from correlation, association, and prediction to experimental determination of causation and molecular mechanisms. Even for some of the most commonly reported intracellular bacteria, like F. nucleatum in CRC, conflicting reports suggest that some strains lack stable gut colonization, coupled with a lack of CRC-promoting effects182,183. Careful evaluation of bacterial engraftment, invasion and downstream impacts on the invaded host, assessed across multiple experimental platforms, may be necessary to draw robust conclusions on an intratumoral or even intracellular microbial causative roles in cancer. Beyond single-species bacterial studies, elucidation of bacterial-bacterial interactions, as well as trans-kingdom interactions with viruses184,185,186, fungi30,187,188 and eukaryotic microorganisms will constitute fascinating topics of research in the coming decade. Progression from the use of cellular systems and animal models to clinical translation will be complex but presents opportunities to harness the gained knowledge in developing novel diagnostics and therapeutics.
In conclusion, some studies suggest that seemingly sterile tumors may harbor a unique low-biomass microbiome, whose bacterial constituents may partially reside within host tumor and immune cells. Future advances in imaging, experimental models, sequencing and data analysis may validate these findings, while accounting for confounders and possible contaminations. Even such definite proofs will only form the basis for the exploration of many unknowns. The rules of engagement by which bacteria populate tumors in specific intratumoral niches remain to be determined. Causal temporal and spatial relations between bacteria and their invaded TME host cells remain elusive, and their downstream effects on tumorigenesis and treatment response are only beginning to be unraveled. The coming decade will likely see a deepening research effort exploring these exciting topics, while exploiting the gained knowledge towards a putative development of novel cancer interventions.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
References
-
Russell, W. An address on a characteristic organism of cancer. Br. Med. J. 2, 1356–1360 (1890).
Google Scholar
-
Butlin, H. T. Malignant tumours and parasitism. Br. Med. J. 1, 45–46 (1884).
Google Scholar
-
Glover, T. Progress in cancer research. Canada Lancet and Practitioner 67, 5 (1926).
-
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Google Scholar
-
ACS. Unproven methods in cancer management: Livingston-Wheeler therapy. CA Cancer J. Clin. 40, 103–108 (1990).
-
Polk, D. B. & Peek, R. M. Jr. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer 10, 403–414 (2010).
Google Scholar
-
Cullin, N., Azevedo Antunes, C., Straussman, R., Stein-Thoeringer, C. K. & Elinav, E. Microbiome and cancer. Cancer Cell 39, 1317–1341 (2021).
Google Scholar
-
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678 (2019).
Google Scholar
-
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019).
Google Scholar
-
Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766 (2014).
Google Scholar
-
Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25, 968–976 (2019).
Google Scholar
-
Kartal, E. et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut 71, 1359–1372 (2022).
Google Scholar
-
Kostic, A. D. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012).
Google Scholar
-
Poore, G. D. et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574 (2020).
Google Scholar
-
Nejman, D. et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020).
Google Scholar
-
Galeano Nino, J. L. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817 (2022).
Google Scholar
-
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Google Scholar
-
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012).
Google Scholar
-
Gihawi, A., Cooper, C. S. & Brewer, D. S. Caution regarding the specificities of pan-cancer microbial structure. Microb. Genom. 9, mgen001088 (2023).
Google Scholar
-
Sepich-Poore, G. D. et al. Reply to: Caution regarding the specificities of pan-cancer microbial structure Preprint at bioRxiv https://doi.org/10.1101/2023.02.10.528049 (2023).
-
Nougayrede, J. P. et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006).
Google Scholar
-
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Google Scholar
-
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Google Scholar
-
Gharaibeh, R. Z. & Jobin, C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut 68, 385–388 (2019).
Google Scholar
-
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e516 (2017).
Google Scholar
-
Geller, L. T. et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017).
Google Scholar
-
Mager, L. F. et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489 (2020).
Google Scholar
-
Choi, Y. et al. Immune checkpoint blockade induces gut microbiota translocation that augments extraintestinal antitumor immunity. Sci. Immunol. 8, eabo2003 (2023).
Google Scholar
-
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–38 (2019).
Google Scholar
-
Saftien, A., Puschhof, J. & Elinav, E. Fungi and cancer. Gut https://doi.org/10.1136/gutjnl-2022-327952 (2023).
-
Mesri, E. A., Feitelson, M. A. & Munger, K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15, 266–282 (2014).
Google Scholar
-
Westermann, A. J., Gorski, S. A. & Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630 (2012).
Google Scholar
-
Easter, Q. T. et al. Polybacterial intracellular coinfection of epithelial stem cells in periodontitis. Preprint at bioRxiv https://doi.org/10.1101/2023.08.23.554343 (2023).
-
Cossart, P. & Sansonetti, P. J. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248 (2004).
Google Scholar
-
Weiss, G. & Schaible, U. E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264, 182–203 (2015).
Google Scholar
-
Aliko, A. et al. Impact of Porphyromonas gingivalis peptidylarginine deiminase on bacterial biofilm formation, epithelial cell invasion, and epithelial cell transcriptional landscape. Sci. Rep. 8, 14144 (2018).
Google Scholar
-
Fattinger, S. A., Sellin, M. E. & Hardt, W. D. Salmonella effector driven invasion of the gut epithelium: breaking in and setting the house on fire. Curr. Opin. Microbiol. 64, 9–18 (2021).
Google Scholar
-
Mempel, M. et al. Invasion of human keratinocytes by Staphylococcus aureus and intracellular bacterial persistence represent haemolysin-independent virulence mechanisms that are followed by features of necrotic and apoptotic keratinocyte cell death. Br. J. Dermatol. 146, 943–951 (2002).
Google Scholar
-
Cróinín, O. T. & Backert, S. Host epithelial cell invasion by Campylobacter jejuni: trigger or zipper mechanism. Front. Cell Infect. Microbiol. 2, 25 (2012).
-
Stelzner, K., Vollmuth, N. & Rudel, T. Intracellular lifestyle of Chlamydia trachomatis and host-pathogen interactions. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-023-00860-y (2023).
-
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Google Scholar
-
Man, S. M., Karki, R. & Kanneganti, T. D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 277, 61–75 (2017).
Google Scholar
-
Aguilar, C., Mano, M. & Eulalio, A. MicroRNAs at the host-bacteria interface: host defense or bacterial offense. Trends Microbiol. 27, 206–218 (2019).
Google Scholar
-
Palmer, G. H., Bankhead, T. & Seifert, H. S. Antigenic variation in bacterial pathogens. Microbiol. Spectr. 4, https://doi.org/10.1128/microbiolspec.VMBF-0005-2015 (2016).
-
Thakur, A., Mikkelsen, H. & Jungersen, G. Intracellular pathogens: host immunity and microbial persistence strategies. J. Immunol. Res. 2019, 1356540 (2019).
Google Scholar
-
Ray, K., Marteyn, B., Sansonetti, P. J. & Tang, C. M. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340 (2009).
Google Scholar
-
Weddle, E. & Agaisse, H. Principles of intracellular bacterial pathogen spread from cell to cell. PLoS Pathog. 14, e1007380 (2018).
Google Scholar
-
Tang, B. et al. MicroRNA-31 induced by Fusobacterium nucleatum infection promotes colorectal cancer tumorigenesis. iScience 26, 106770 (2023).
Google Scholar
-
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Google Scholar
-
Casasanta, M. A. et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 13, https://doi.org/10.1126/scisignal.aba9157 (2020).
-
Brennan, C. A. & Garrett, W. S. Fusobacterium nucleatum – symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166 (2019).
Google Scholar
-
Ghaddar, B. et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell 40, 1240–1253.e1245 (2022).
Google Scholar
-
Huang, S. T. et al. Intratumoral levels and prognostic significance of Fusobacterium nucleatum in cervical carcinoma. Aging 12, 23337–23350 (2020).
Google Scholar
-
Yamamura, K. et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 22, 5574–5581 (2016).
Google Scholar
-
Yamamura, K. et al. Intratumoral Fusobacterium nucleatum levels predict therapeutic response to neoadjuvant chemotherapy in esophageal squamous cell carcinoma. Clin. Cancer Res. 25, 6170–6179 (2019).
Google Scholar
-
Wang, X. et al. Porphyromonas gingivalis promotes colorectal carcinoma by activating the hematopoietic NLRP3 inflammasome. Cancer Res. 81, 2745–2759 (2021).
Google Scholar
-
Lamont, R. J., Fitzsimonds, Z. R., Wang, H. & Gao, S. Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontology 89, 154–165 (2022).
-
Fan, X. et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67, 120–127 (2018).
Google Scholar
-
Okumura, S. et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 12, 5674 (2021).
Google Scholar
-
Lo, C. H. et al. Enrichment of Prevotella intermedia in human colorectal cancer and its additive effects with Fusobacterium nucleatum on the malignant transformation of colorectal adenomas. J. Biomed. Sci. 29, 88 (2022).
Google Scholar
-
Gursoy, U. K., Kononen, E. & Uitto, V. J. Prevotella intermedia ATCC 25611 targets host cell lamellipodia in epithelial cell adhesion and invasion. Oral. Microbiol. Immunol. 24, 304–309 (2009).
Google Scholar
-
Pushalkar, S. et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 8, 403–416 (2018).
Google Scholar
-
Fletcher, A. A., Kelly, M. S., Eckhoff, A. M. & Allen, P. J. Revisiting the intrinsic mycobiome in pancreatic cancer. Nature 620, E1–E6 (2023).
Google Scholar
-
Dohlman, A. B. et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe https://doi.org/10.1016/j.chom.2020.12.001 (2021).
-
Fuks, G. et al. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 6, 17 (2018).
Google Scholar
-
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Google Scholar
-
de Goffau, M. C. et al. Recognizing the reagent microbiome. Nat. Microbiol. 3, 851–853 (2018).
Google Scholar
-
Austin, G. I. et al. Contamination source modeling with SCRuB improves cancer phenotype prediction from microbiome data. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01696-w (2023).
-
Westermann, A. J. & Vogel, J. Cross-species RNA-seq for deciphering host-microbe interactions. Nat. Rev. Genet. 22, 361–378 (2021).
Google Scholar
-
Seelbinder, B. et al. Triple RNA-seq reveals synergy in a human virus-fungus co-infection model. Cell Rep. 33, 108389 (2020).
Google Scholar
-
Bullman, S. INVADEseq to study the intratumoural microbiota at host single-cell resolution. Nat. Rev. Cancer 23, 189 (2023).
Google Scholar
-
Guven-Maiorov, E. et al. HMI-PRED: a web server for structural prediction of host-microbe interactions based on interface mimicry. J. Mol. Biol. 432, 3395–3403 (2020).
Google Scholar
-
Weiner, A. & Enninga, J. The pathogen-host interface in three dimensions: correlative FIB/SEM applications. Trends Microbiol 27, 426–439 (2019).
Google Scholar
-
Strauss, J. et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978 (2011).
Google Scholar
-
Ponath, F., Zhu, Y., Cosi, V. & Vogel, J. Expanding the genetic toolkit helps dissect a global stress response in the early-branching species Fusobacterium nucleatum. Proc. Natl. Acad. Sci. USA 119, e2201460119 (2022).
Google Scholar
-
Puschhof, J. et al. Intestinal organoid cocultures with microbes. Nat. Protoc. 16, 4633–4649 (2021).
Google Scholar
-
Xue, C. et al. Current understanding of the intratumoral microbiome in various tumors. Cell Rep. Med. 4, 100884 (2023).
Google Scholar
-
Guiney, D. G., Hasegawa, P. & Cole, S. P. Helicobacter pylori preferentially induces interleukin 12 (IL-12) rather than IL-6 or IL-10 in human dendritic cells. Infect. Immun. 71, 4163–4166 (2003).
Google Scholar
-
Hafsi, N. et al. Human dendritic cells respond to Helicobacter pylori, promoting NK cell and Th1-effector responses in vitro. J. Immunol. 173, 1249–1257 (2004).
Google Scholar
-
Bagheri, N., Salimzadeh, L. & Shirzad, H. The role of T helper 1-cell response in Helicobacter pylori-infection. Micro. Pathog. 123, 1–8 (2018).
Google Scholar
-
Sigal, M. et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature 548, 451–455 (2017).
Google Scholar
-
Correa, P. & Piazuelo, M. B. The gastric precancerous cascade. J. Dig. Dis. 13, 2–9 (2012).
Google Scholar
-
Pan, G. et al. Helicobacter pylori promotes gastric cancer progression by upregulating semaphorin 5A expression via ERK/MMP9 signaling. Mol. Ther. Oncolytics 22, 256–264 (2021).
Google Scholar
-
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Google Scholar
-
Castellarin, M. et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. https://doi.org/10.1101/gr.126516.111 (2011).
-
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013).
Google Scholar
-
Liu, H. et al. Fusobacterium nucleatum exacerbates colitis by damaging epithelial barriers and inducing aberrant inflammation. J. Dig. Dis. 21, 385–398 (2020).
Google Scholar
-
Gursoy, U. K., Kononen, E. & Uitto, V. J. Intracellular replication of Fusobacteria requires new actin filament formation of epithelial cells. APMIS 116, 1063–1070 (2008).
Google Scholar
-
Dorn, B. R., Burks, J. N., Seifert, K. N. & Progulske-Fox, A. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol Lett. 187, 139–144 (2000).
Google Scholar
-
Swidsinski, A. et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 115, 281–286 (1998).
Google Scholar
-
Raisch, J., Rolhion, N., Dubois, A., Darfeuille-Michaud, A. & Bringer, M. A. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab. Investig. 95, 296–307 (2015).
Google Scholar
-
Jans, M. et al. Colibactin-induced genotoxicity and colorectal cancer exacerbation critically depends on adhesin-mediated epithelial binding. Preprint at bioRxiv https://doi.org/10.1101/2023.08.16.553526 (2023).
-
Cano, C. E. et al. Homotypic cell cannibalism, a cell-death process regulated by the nuclear protein 1, opposes to metastasis in pancreatic cancer. EMBO Mol. Med. 4, 964–979 (2012).
Google Scholar
-
Chakladar, J. et al. The pancreatic microbiome is associated with carcinogenesis and worse prognosis in males and smokers. Cancers 12, https://doi.org/10.3390/cancers12092672 (2020).
-
Badger, J. L., Stins, M. F. & Kim, K. S. Citrobacter freundii invades and replicates in human brain microvascular endothelial cells. Infect. Immun. 67, 4208–4215 (1999).
Google Scholar
-
Torraca, V., Holt, K. & Mostowy, S. Shigella sonnei. Trends Microbiol. 28, 696–697 (2020).
Google Scholar
-
Petersen, A. M. & Krogfelt, K. A. Helicobacter pylori: an invading microorganism? A review. FEMS Immunol. Med. Microbiol 36, 117–126 (2003).
Google Scholar
-
Trikudanathan, G., Philip, A., Dasanu, C. A. & Baker, W. L. Association between Helicobacter pylori infection and pancreatic cancer. A cumulative meta-analysis. JOP 12, 26–31 (2011).
Google Scholar
-
Aykut, B. et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267 (2019).
Google Scholar
-
Dohlman, A. B. et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell 185, 3807–3822.e3812 (2022).
Google Scholar
-
Urbaniak, C. et al. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 82, 5039–5048 (2016).
Google Scholar
-
Fu, A. et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185, 1356–1372.e1326 (2022).
Google Scholar
-
Shanmughapriya, S. et al. Viral and bacterial aetiologies of epithelial ovarian cancer. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2311–2317 (2012).
Google Scholar
-
Paavonen, J., Turzanski Fortner, R., Lehtinen, M. & Idahl, A. Chlamydia trachomatis, pelvic inflammatory disease, and epithelial ovarian cancer. J. Infect. Dis. 224, S121–S127 (2021).
Google Scholar
-
Yang, X. et al. Chlamydia trachomatis infection: their potential implication in the etiology of cervical cancer. J. Cancer 12, 4891–4900 (2021).
Google Scholar
-
Siegl, C. & Rudel, T. Modulation of p53 during bacterial infections. Nat. Rev. Microbiol. 13, 741–748 (2015).
Google Scholar
-
Fischer, A. & Rudel, T. Subversion of cell-autonomous host defense by Chlamydia infection. Curr. Top. Microbiol. Immunol. 412, 81–106 (2018).
Google Scholar
-
Wong, L. M. et al. Comparative analysis of age- and gender-associated microbiome in lung adenocarcinoma and lung squamous cell carcinoma. Cancers 12, https://doi.org/10.3390/cancers12061447 (2020).
-
Apostolou, P. et al. Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg. 6, 137 (2011).
Google Scholar
-
Chai, X. et al. Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut Microbes 15, 2156255 (2023).
Google Scholar
-
Iida, N. et al. Chronic liver disease enables gut Enterococcus faecalis colonization to promote liver carcinogenesis. Nat. Cancer 2, 1039–1054 (2021).
Google Scholar
-
Nunez, N. et al. The unforeseen intracellular lifestyle of Enterococcus faecalis in hepatocytes. Gut Microbes 14, 2058851 (2022).
Google Scholar
-
Huang, Y. et al. Identification of Helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J. Clin. Pathol. 57, 1273–1277 (2004).
Google Scholar
-
Kumar, N. G. et al. Pseudomonas aeruginosa can diversify after host cell invasion to establish multiple intracellular niches. mBio 13, e0274222 (2022).
Google Scholar
-
Al-Hebshi, N. N. et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 7, 1834 (2017).
Google Scholar
-
Ma, J. et al. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers 12, https://doi.org/10.3390/cancers12092524 (2020).
-
Kalaora, S. et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138–143 (2021).
Google Scholar
-
Konishi, Y. et al. Development and evaluation of a colorectal cancer screening method using machine learning-based gut microbiota analysis. Cancer Med. 11, 3194–3206 (2022).
Google Scholar
-
Scott, N., Whittle, E., Jeraldo, P. & Chia, N. A systemic review of the role of enterotoxic Bacteroides fragilis in colorectal cancer. Neoplasia 29, 100797 (2022).
Google Scholar
-
Lamont, R. J. & Hajishengallis, G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183 (2015).
Google Scholar
-
Darveau, R. P., Belton, C. M., Reife, R. A. & Lamont, R. J. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66, 1660–1665 (1998).
Google Scholar
-
Saito, A. et al. Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts. Micro. Pathog. 53, 234–242 (2012).
Google Scholar
-
Ribet, D. & Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 17, 173–183 (2015).
Google Scholar
-
Siggins, M. K. et al. Extracellular bacterial lymphatic metastasis drives Streptococcus pyogenes systemic infection. Nat. Commun. 11, 4697 (2020).
Google Scholar
-
Heikkila, P., But, A., Sorsa, T. & Haukka, J. Periodontitis and cancer mortality: register-based cohort study of 68,273 adults in 10-year follow-up. Int. J. Cancer 142, 2244–2253 (2018).
Google Scholar
-
Michaud, D. S., Fu, Z., Shi, J. & Chung, M. Periodontal disease, tooth loss, and cancer risk. Epidemiol. Rev. 39, 49–58 (2017).
Google Scholar
-
Han, Y. W. et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68, 3140–3146 (2000).
Google Scholar
-
Gnanasekaran, J. et al. Intracellular Porphyromonas gingivalis promotes the tumorigenic behavior of pancreatic carcinoma cells. Cancers 12, https://doi.org/10.3390/cancers12082331 (2020).
-
Abed, J. et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20, 215–225 (2016).
Google Scholar
-
Nakagaki, H. et al. Fusobacterium nucleatum envelope protein FomA is immunogenic and binds to the salivary statherin-derived peptide. Infect. Immun. 78, 1185–1192 (2010).
Google Scholar
-
Riquelme, E. et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e712 (2019).
Google Scholar
-
Huang, J. & Huang, J. Microbial biomarkers for lung cancer: current understandings and limitations. J. Clin. Med. 11, https://doi.org/10.3390/jcm11247298 (2022).
-
Yang, M. et al. Bacteria-mediated cancer therapies: opportunities and challenges. Biomater. Sci. 9, 5732–5744 (2021).
Google Scholar
-
Chen, Y., Wu, F. H., Wu, P. Q., Xing, H. Y. & Ma, T. The role of the tumor microbiome in tumor development and its treatment. Front. Immunol. 13, 935846 (2022).
Google Scholar
-
Bertocchi, A. et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell 39, 708–724.e711 (2021).
Google Scholar
-
Mandell, G. L. Interaction of intraleukocytic bacteria and antibiotics. J. Clin. Investig. 52, 1673–1679 (1973).
Google Scholar
-
Vaudaux, P. & Waldvogel, F. A. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob. Agents Chemother. 16, 743–749 (1979).
Google Scholar
-
Gur, C. et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42, 344–355 (2015).
Google Scholar
-
Yang, Y. et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-Like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology 152, 851–866.e824 (2017).
Google Scholar
-
Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 20, https://doi.org/10.15252/embr.201847638 (2019).
-
Elsinghorst, E. A. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236, 405–420 (1994).
Google Scholar
-
Kim, R. et al. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 12, 015006 (2019).
Google Scholar
-
Sasaki, N. et al. Development of a scalable coculture system for gut anaerobes and human colon epithelium. Gastroenterology 159, 388–390.e385 (2020).
Google Scholar
-
Puschhof, J., Pleguezuelos-Manzano, C. & Clevers, H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 29, 867–878 (2021).
Google Scholar
-
Kasper, S. H. et al. Colorectal cancer-associated anaerobic bacteria proliferate in tumor spheroids and alter the microenvironment. Sci. Rep. 10, 5321 (2020).
Google Scholar
-
Xu, B., Zhou, M., Qiu, W., Ye, J. & Feng, Q. CCR7 mediates human breast cancer cell invasion, migration by inducing epithelial-mesenchymal transition and suppressing apoptosis through AKT pathway. Cancer Med. 6, 1062–1071 (2017).
Google Scholar
-
Jalili-Firoozinezhad, S. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531 (2019).
Google Scholar
-
Sontheimer-Phelps, A. et al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell Mol. Gastroenterol. Hepatol. 9, 507–526 (2020).
Google Scholar
-
Grassart, A. et al. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting shigella infection. Cell Host Microbe 26, 435–444.e434 (2019).
Google Scholar
-
Avraham, R. et al. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell 162, 1309–1321 (2015).
Google Scholar
-
Lee, P. & Tan, K. S. Fusobacterium nucleatum activates the immune response through retinoic acid-inducible gene I. J. Dent. Res. 93, 162–168 (2014).
Google Scholar
-
Zhang, Y. et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-kappaB/ICAM1 axis. Gut Microbes 14, 2038852 (2022).
Google Scholar
-
Yu, T. C. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e516 (2017).
Google Scholar
-
Qian, B. Z. & Pollard, J. W. Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010).
Google Scholar
-
Kaplan, C. W. et al. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J. Dent. Res. 84, 700–704 (2005).
Google Scholar
-
Kaplan, C. W. et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect. Immun. 78, 4773–4778 (2010).
Google Scholar
-
Naghavian, R. et al. Microbial peptides activate tumour-infiltrating lymphocytes in glioblastoma. Nature https://doi.org/10.1038/s41586-023-06081-w (2023).
-
Yang, J. et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020).
Google Scholar
-
Kong, C. et al. Fusobacterium nucleatum promotes the development of colorectal cancer by activating a cytochrome P450/epoxyoctadecenoic acid axis via TLR4/Keap1/NRF2 signaling. Cancer Res. 81, 4485–4498 (2021).
Google Scholar
-
Lu, X. et al. Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. 40, 111127 (2022).
Google Scholar
-
Ternes, D. et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat. Metab. 4, 458–475 (2022).
Google Scholar
-
Chen, S. et al. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes 11, 511–525 (2020).
Google Scholar
-
Guo, S. et al. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut https://doi.org/10.1136/gutjnl-2020-321187 (2020).
-
Alimonti, A. et al. New approaches to prevent intestinal toxicity of irinotecan-based regimens. Cancer Treat. Rev. 30, 555–562 (2004).
Google Scholar
-
Lehouritis, P. et al. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 5, 14554 (2015).
Google Scholar
-
LaCourse, K. D. et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. 41, 111625 (2022).
Google Scholar
-
Serna, G. et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 31, 1366–1375 (2020).
Google Scholar
-
Harish, B. N. & Menezes, G. A. Determination of antimicrobial resistance in Salmonella spp. Methods Mol. Biol. 1225, 47–61 (2015).
Google Scholar
-
Buccini, D. F., Cardoso, M. H. & Franco, O. L. Antimicrobial peptides and cell-penetrating peptides for treating intracellular bacterial infections. Front. Cell Infect. Microbiol. 10, 612931 (2020).
Google Scholar
-
Gomarasca, M. et al. Bacterium-derived cell-penetrating peptides deliver gentamicin to kill intracellular pathogens. Antimicrob. Agents Chemother. 61, https://doi.org/10.1128/AAC.02545-16 (2017).
-
Singh, R., Patil, S., Singh, N. & Gupta, S. Dual functionality nanobioconjugates targeting intracellular bacteria in cancer cells with enhanced antimicrobial activity. Sci. Rep. 7, 5792 (2017).
Google Scholar
-
Zheng, D. W. et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 3, 717–728 (2019).
Google Scholar
-
Gogokhia, L. et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe 25, 285–299.e288 (2019).
Google Scholar
-
Federici, S. et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 185, 2879–2898.e2824 (2022).
Google Scholar
-
Raman, V. et al. Intracellular delivery of protein drugs with an autonomously lysing bacterial system reduces tumor growth and metastases. Nat. Commun. 12, 6116 (2021).
Google Scholar
-
Costa, S. C., Girard, P. A., Brehelin, M. & Zumbihl, R. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect. Immun. 77, 1022–1030 (2009).
Google Scholar
-
Wang, M. et al. The roles of two type VI secretion systems in Cronobacter sakazakii ATCC 12868. Front. Microbiol. 9, 2499 (2018).
Google Scholar
-
Kreitz, J. et al. Programmable protein delivery with a bacterial contractile injection system. Nature 616, 357–364 (2023).
Google Scholar
-
Avogadri, F. et al. Cancer immunotherapy based on killing of Salmonella-infected tumor cells. Cancer Res. 65, 3920–3927 (2005).
Google Scholar
-
Gihawi, A. et al. Major data analysis errors invalidate cancer microbiome findings. Preprint at bioRxiv https://doi.org/10.1101/2023.07.28.550993 (2023).
-
Kennedy, K. M. et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature 613, 639–649 (2023).
Google Scholar
-
Queen, J. et al. Comparative analysis of colon cancer-derived Fusobacterium nucleatum subspecies: inflammation and colon tumorigenesis in murine models. mBio 13, e0299121 (2021).
Google Scholar
-
Tomkovich, S. et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 77, 2620–2632 (2017).
Google Scholar
-
Chin, V. K. et al. Mycobiome in the gut: a multiperspective review. Mediators Inflamm. 2020, 9560684 (2020).
Google Scholar
-
Ogunrinola, G. A., Oyewale, J. O., Oshamika, O. O. & Olasehinde, G. I. The human microbiome and its impacts on health. Int. J. Microbiol. 2020, 8045646 (2020).
Google Scholar
-
Krump, N. A. & You, J. Molecular mechanisms of viral oncogenesis in humans. Nat. Rev. Microbiol. 16, 684–698 (2018).
Google Scholar
-
Dohlman, A. B. et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe 29, 281–298.e285 (2021).
Google Scholar
-
Narunsky-Haziza, L. et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 185, 3789–3806.e3717 (2022).
Google Scholar
Acknowledgements
We thank Tali Wiesel, Weizmann Institute of Science, for graphical help with figure preparation. L.S. is supported by the Helmholtz Foundation. E.E. is supported by the Leona M. and Harry B. Helmsley Charitable Trust; Adelis Foundation; Ben B. And Joyce E. Eisenberg Foundation; Estate of Bernard Bishin for the WIS-Clalit Program; Jeanne and Joseph Nissim Center for Life Sciences Research; Miel de Botton; Swiss Society Institute for Cancer Prevention Research; Belle S. and Irving E. Meller Center for the Biology of Aging; Sagol Institute for Longevity Research; Sagol Weizmann-MIT Bridge Program; Norman E. Alexander Family M Foundation Coronavirus Research Fund; Mike and Valeria Rosenbloom Foundation; Daniel Morris Trust; Isidore and Penny Myers Foundation; Vainboim Family; and by grants funded by the European Research Council; Israel Science Foundation; Israel Ministry of Science and Technology; Israel Ministry of Health; the German-Israeli Helmholtz International Research School: Cancer-TRAX (HIRS-0003); Helmholtz Association’s Initiative and Networking Fund; Minerva Foundation; Garvan Institute; European Crohn’s and Colitis Organization; Deutsch-Israelische Projektkooperation; IDSA Foundation; WIS-MIT grant; Emulate; Charlie Teo Foundation; Mark Foundation for Cancer Research, and Welcome Trust. E.E. is the incumbent of Sir Marc and Lady Tania Feldmann Professorial Chair of Immunology; a senior fellow, Canadian Institute of Advanced Research; and an international scholar, The Bill & Melinda Gates Foundation and Howard Hughes Medical Institute. J.P. is supported by the Helmholtz Foundation, the Hector Foundation, and Emulate Inc.
Author information
Authors and Affiliations
Contributions
L.S. and M.M. are co-first authors. L.S. and M.M. conceptualized and wrote the manuscript. E.E. and J.P. wrote the manuscript and supervised the review.
Corresponding authors
Ethics declarations
Competing interests
E.E. is a scientific founder of Daytwo and BiomX, and a paid consultant to Igen, PurposeBio, Aposense, and Zoe. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Reporting Summary
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Schorr, L., Mathies, M., Elinav, E. et al. Intracellular bacteria in cancer—prospects and debates.
npj Biofilms Microbiomes 9, 76 (2023). https://doi.org/10.1038/s41522-023-00446-9
-
Received: 13 June 2023
-
Accepted: 26 September 2023
-
Published: 09 October 2023
-
DOI: https://doi.org/10.1038/s41522-023-00446-9