Infection
Herpes Virus Infections in Kidney Transplant Patients (HINT) – a prospective observational cohort study
The Herpes Virus Infections in Kidney Transplant Patients (HINT) study is a prospective observational cohort study. Kidney transplant recipients transplanted after January 1, 2019, and kidney transplant candidates on the waiting list for kidney transplantation at Copenhagen University Hospital – Rigshospitalet, Odense University Hospital, or Aarhus University Hospital who are offered a RZV vaccine will be invited to participate in the study until February 1, 2025. Recruitment of participants started January 16, 2023 and is currently ongoing.
The kidney transplant centers at Copenhagen University Hospital – Rigshospitalet, Odense University Hospital, or Aarhus University Hospital are responsible for the inclusion and follow-up of participants, while researchers from the department of health technology at Technical University of Denmark (DTU) are responsible for the immunological analyses. The study is performed in collaboration with the herpesvirus immunology in solid organ transplant recipients – liver transplant study (HISTORY) and parts of the methodology follows what has previously been described in the HISTORY study protocol [21]. A STROBE [22] checklist is provided as supplementary material (supplementary material).
Vaccinations
All participants who accept the offer to receive RZV vaccination are vaccinated according to Danish guidelines at their local transplant center. Therefore, vaccinations are performed uniformly at all three centers by administering two doses of vaccine with two months between each dose. All vaccinations administered are a part of routine clinical care.
Eligibility criteria
Participants must be either on the waiting list for kidney transplantation OR first time transplanted after January 1, 2019, AND above 18 years old, able to provide informed consent, and have been offered a RZV vaccine. Patients are eligible to participate whether they accept RZV vaccination or not.
Blood sampling
Venous blood samples are collected from participants at scheduled visits to the clinical sites. All participants will provide a blood sample at inclusion i.e., at the time they are offered the first dose of RZV vaccination. Hereafter, all participants will be invited for repeated blood sampling 1, 2, 6, and 12 months after the first dose of RZV was offered. Furthermore, participants who are on the waiting list for transplantation and transplanted during the study will be invited for blood sampling at 6 and 12 months after transplantation. If a participant develops herpes zoster during the study period, they will be invited for blood sampling immediately after the infection has been diagnosed. At each sample time, 60 mL of venous blood will be collected in lithium-heparin and EDTA-coated collection tubes.
Within 4 h after collection, blood collected in lithium-heparin-coated collection blood tubes will be transported to the laboratory for purification of peripheral blood mononuclear cells (PBMC) using density gradient centrifugation. EDTA-coated collection tubes will be stored on ice immediately after collection and centrifuged within 1 h, after which plasma will be separated and stored at -80 °C. After primary processing at either Copenhagen University Hospital – Rigshospitalet, Odense University Hospital, or Aarhus University Hospital, all PBMC and plasma samples will be stored in liquid nitrogen at a biobank at the Department of Health Technology, DTU.
Questionnaire
At inclusion, participants will be asked to fill out a questionnaire about health, lifestyle, socioeconomic status, current and previous use of medication and vaccinations. The questionnaire will be entered directly into REDCap [23]—a secured digital platform or on paper (supplementary material). At each following study visit, participants will be asked to fill out a questionnaire regarding changes related to transplantation, vaccinations, rejection episodes, and infections since the last study visit (supplementary material).
Antibody assay
The anti-glycoprotein E antibody response rate will be determined by Enzyme-Linked Immunosorbent Assay (ELISA). Glycoprotein E is the main target for VZV-specific immune responses [24]. We will establish and validate a previously reported ELISA and measure anti-glycoprotein E antibody concentrations in plasma samples collected from all participants at all time points. The anti-glycoprotein E antibody response rate will be defined as the percentage of subjects with anti-glycoprotein E antibody concentration ≥ 4-fold higher than at baseline.
Identification of VZV specific T cells
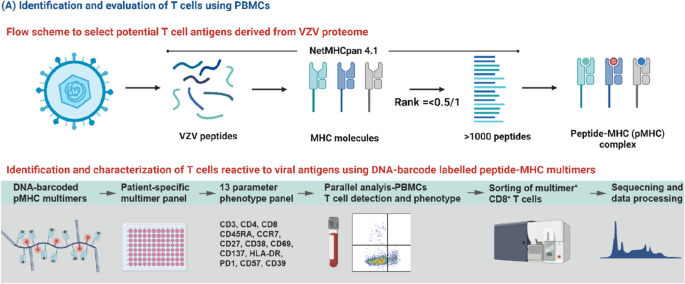
To identify the complete repertoire of VZV-specific T cells and their immunodominance towards VZV, we will perform a broad T cell screening towards potential T cell epitopes in VZV, as previously described [21]. Human leukocyte antigen (HLA)-class I presented epitopes will be predicted (NetMHCpan-4.1) and synthetized to make peptide-HLA multimers matching the HLA profile of the individual participant (Fig. 1). We will cover the 12 most frequent HLA haplotypes [25]. Using DNA-barcode labeled peptide-major histocompatibility complex (pMHC) multimers and subsequent flow cytometry-based sorting of VZV-specific T cells, we can determine the T cell recognition to a large library of potential epitopes (> 1000/sample) [26, 27]. The sequential sampling pre- and post-transplantation will allow a comparative assessment and monitoring of T cell frequency, phenotype and fitness at different levels of immunosuppression.
Overview of methods for identification and evaluation of T cells using peripheral blood mononuclear cells
Using the NetMHCpan 4.1 platform, as previously described in the HISTORY study protocol [21], potential varicella-zoster virus (VZV) peptide-MHC (pMHC) complexes will be predicted corresponding to the most frequent HLA haplotypes amongst participants. The pMHC complexes will be combined in a DNA-barcoded patient-specific pMHC multimer. Peripheral blood mononuclear cells (PBMCs) will be examined for their detection of pMHC multimers and sorted using flow cytometry based on their expression of phenotypic cell surface markers. The reactive T cells may be further examined for their frequency, T cell receptor sequence, and potential for ex vivo expansion [21].
Phenotype and cytokine profiling
Phenotyping and functional characterization of specific T cell populations towards immunodominant epitopes in VZV will be carried out, as previously described in the HISTORY study protocol [21], by using a flow cytometry panel of 13 cell surface and intracellular markers of activation, senescence, and exhaustion (Fig. 1). Furthermore, we will measure the cytokine profiles in plasma samples using an established Luminex MILLIPLEX assay to simultaneously quantify 41 analytes that map the complete cytokine signature including interleukins, chemokines, and growth factors (Fig. 2).

Overview of methods for cytokine profiling using Luminex MILLIPLEX® assay
Cytokine profiling will be performed as previously described in the HISTORY study protocol [21]. Plasma contains an unspecified concentration of analytes of interest, i.e. cytokines, chemokines, interleukins and growth factors. To determine the concentration of the analytes, each Luminex well contains multiple types of color-coded beads conjugated to a capture antibody that binds the specific analyte of interest. In presence of the analyte, a biotin-conjugated detection antibody will bind to the capture antibody-analyte complex. Adding phycoerythrin-labeled streptavidin will produce fluorescence that is color-specific for each analyte and allows for measurement of the mean fluorescence intensity (MFI) of each analyte in each Luminex well. Comparing MFI at different dilution levels with standard solutions allows for determination of the concentration of the analytes of interest.
T cell receptor sequencing
We will apply a technology that allows for large-scale collection of T cell receptor (TCR) sequences paired to different peptide targets based on their pMHC recognition motif through a pMHC multimer linked to a DNA barcode [28]. Using single-cell capture and analysis systems based on sorted pMHC responsive T cells, we can pair the TCR sequences and assign the pMHC specificity based on the co-attached DNA barcode. Using this platform, we will identify T cell clonotypes of relevance for VZV recognition and describe their functional characteristics. We will implement parallel phenotyping of immune cells using a 150 + marker panel of DNA barcoded antibodies that target a broad array of lineage markers, activation markers, exhaustion markers, and regulatory markers.
Clinical data
Data on demographics, comorbidities, biochemistry, medication, allograft rejection, pathology, HLA, and hospital admissions will be collected from electronical hospital records. All Danish hospital records since 1 December 2010 are accessible through a nationwide digital platform. Data on vaccinations will be collected through the Danish vaccination register (DDV) [29]. Since 2015, it has been mandatory to register all vaccinations administered in Denmark in DDV. Data on infections will be collected from the Danish Microbiology database (MiBa) [30], which is a nationwide database including all microbiological samples collected at both hospitals and general practitioners in Denmark.
Statistics
Sample size
The purpose of the study is to describe the T-cell and anti-glycoprotein E antibody response to RZV and herpes zoster in kidney transplant candidates and recipients and to explore factors for immune non-response at different times before and after transplantation. In Denmark about 390 patients are active on the waiting list for kidney transplantation and about 250 kidney transplantations are performed each year [31]. We aim to include 250 kidney transplant recipients who are 6–12 months from transplantation, 125 who are 12–24 months from transplantation, and 125 who are > 24 months. Furthermore, since some of the transplant candidates may not be transplanted during the study period, we will include 375 patients who are on the waiting list. Given the descriptive and exploratory scope of the study and lack of existing data regarding outcomes of interest, a formal sample size calculation cannot be conducted. The study is not powered for clinical efficacy estimates.
Statistical analyses
Continuous data will be compared using Student’s t test or Mann–Whitney U test, and categorical data will be compared using Pearson’s χ2 or Fisher’s exact tests, as appropriate. Mixed linear models will be used to evaluate the development of antibody and T cell immunity towards VZV over time and to explore risk factors for non-response.