Congenital disorders
Altered Gut Microbiota and Short-chain Fatty Acids in Chinese Children with Constipated Autism Spectrum Disorder
Abstract
Gastrointestinal symptoms are more prevalent in children with autism spectrum disorder (ASD) than in typically developing (TD) children. Constipation is a significant gastrointestinal comorbidity of ASD, but the associations among constipated autism spectrum disorder (C-ASD), microbiota and short-chain fatty acids (SCFAs) are still debated. We enrolled 80 children, divided into the C-ASD group (n = 40) and the TD group (n = 40). In this study, an integrated 16S rRNA gene sequencing and gas chromatography–mass spectrometry-based metabolomics approach was applied to explore the association of the gut microbiota and SCFAs in C-ASD children in China. The community diversity estimated by the Observe, Chao1, and ACE indices was significantly lower in the C-ASD group than in the TD group. We observed that Ruminococcaceae_UCG_002, Erysipelotrichaceae_UCG_003, Phascolarctobacterium, Megamonas, Ruminiclostridium_5, Parabacteroides, Prevotella_2, Fusobacterium, and Prevotella_9 were enriched in the C-ASD group, and Anaerostipes, Lactobacillus, Ruminococcus_gnavus_group, Lachnospiraceae_NK4A136_group, Ralstonia, Eubacterium_eligens_group, and Ruminococcus_1 were enriched in the TD group. The propionate levels, which were higher in the C-ASD group, were negatively correlated with the abundance of Lactobacillus taxa, but were positively correlated with the severity of ASD symptoms. The random forest model, based on the 16 representative discriminant genera, achieved a high accuracy (AUC = 0.924). In conclusion, we found that C-ASD is related to altered gut microbiota and SCFAs, especially decreased abundance of Lactobacillus and excessive propionate in faeces, which provide new clues to understand C-ASD and biomarkers for the diagnosis and potential strategies for treatment of the disorder. This study was registered in the Chinese Clinical Trial Registry (www.chictr.org.cn; trial registration number ChiCTR2100052106; date of registration: October 17, 2021).
Introduction
Autism spectrum disorder (ASD) is a category of complex neurodevelopmental disorders characterized by diminished social skills, communication impairments, and restricted interests or repetitive behaviours1. ASD affects over 5 million Americans, with a prevalence of approximately 2.3% in children, and the prevalence of ASD in boys is 4.2 times higher than that in girls2. The incidence of ASD in China has also increased in recent years. According to a study, the incidence of ASD in China increased by approximately 3.9% in 20183.
Many children with ASD experience co-occurring conditions, such as gastrointestinal (GI) problems and picky eating4,5. A comprehensive meta-analysis showed that children with ASD were more than 4 times as likely to have GI disorders as children without ASD, with constipation, diarrhoea, and abdominal pain being the most common symptoms6. Constipation is an important GI comorbidity of ASD, and the incidence of constipation increases with greater social impairments and reduced language skills4. The median prevalence of ASD patients with one or more GI symptoms is 46.8%7. The relationship between internalizing symptoms and GI problems in youth with ASD is bidirectional8. Moreover, improvement in GI symptoms led to a decrease in the severity of social and emotional disorders in constipated autism spectrum disorder (C-ASD) children9.The bidirectional interaction between the gut microbiota and the brain is known as the “microbiota-gut-brain axis10,11,12,13”. Changes in the gut microbiota and metabolite composition have been found in both human ASD patients and animal models of ASD14,15. Children with ASD have an abnormal gut microbiota composition compared with typically developing (TD) children16,17,18,19,20, and studies have also found higher levels of certain pathogenic bacterial species in children with ASD than in normal children21. Clostridia, Bacteroidetes, and Dorea significantly increased in ASD, producing metabolites including short-chain fatty acids (SCFAs) such as propionate (PPA) and butyrate19. Previous studies have mainly focused on altered metabolites and gut microbiota of ASD patients22,23 or on altered gut microbiota of C-ASD24. A study by Wong et al. did not recruited girls25, which compared the microbiome of ASD with and without GI symptoms. However, few studies have focused on SCFAs in the faeces of C-ASD. A study by Alshammari et al. found that the highest incidence of Clostridium perfringens, a PPA-related bacteria, was found in the C-ASD group26. While numerous studies have highlighted differences in gut microbiota between individuals with ASD and TD, the search for beneficial bacteria or viable metabolic products to clinically treat ASD has been unsuccessful. This complex array of factors influencing gut microbial communities may stem partly from the interaction between culture, diet, environment, and customs constitutes. However, there is currently insufficient information available regarding the composition of gut microbiota and SCFAs in children with C-ASD in southern China.
In this study, we aimed to clarify the differences in faecal bacterial diversity and SCFAs levels in C-ASD children and investigate the association of the gut microbiota and SCFAs with C-ASD clinical parameters. Based on the representative discriminant model of dominant genera, we aimed to identify a universal set of microbial biomarkers to predict C-ASD.
Results
Characteristics and clinical indices of the participants
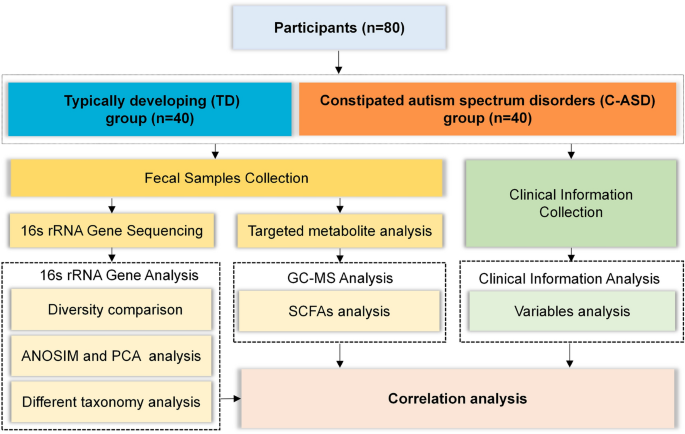
After application of the inclusion and exclusion criteria, 40 TD children and 40 children with C-ASD were enrolled, and their data were analysed (Fig. 1). The patients’ clinical characteristics (including maternal age, gestational age, delivery mode, feeding patterns, maternal educational level, maternal smoking history, maternal drinking history, paternal educational level, paternal smoking history, and paternal drinking history) were compared between the TD and C-ASD groups. Additionally, we compared age, sex, height, weight, body mass index (BMI), birth weight, and Children’s Eating Behaviour Questionnaire (CEBQ) score between the TD group and the C-ASD group. There were no differences in all comparisons shown in Table 1. In the CEBQ scores, food fussiness, satiety responsiveness and desire to drink subscale scores were notably higher in the C-ASD group (Appendix 2. Table A1).
Flow diagram of this study.
Comparison of the diversity of the gut microbiota between the TD and C-ASD groups
The alpha diversity estimated by the Observe, Chao1, and ACE indices was significantly lower in the C-ASD group than in the TD group (p < 0.05). There was no statistically significant difference between the TD group and the C-ASD group in the J index, Simpson index, or Shannon index (p > 0.05) (Fig. 2A). Analysis of similarities (ANOSIM) was applied to compare within- and between-group similarity through a distance measure to evaluate the null hypothesis that the average grade resemblance between samples within a group aligned with the average rank similarity between samples belonging to different groups. There were marked differences in the population distribution between the TD group and the C-ASD group (Fig. 2B). PCoA was performed by using the distance matrix calculated from the species composition of the sample. The x axis and the y axis represent the contribution of the first principal component (PCoA1; 17.66%) and the second principal component (PCoA2; 7.82%), respectively (p = 0.001) (Fig. 2C). The Venn diagram indicates the shared and unique OTUs of the gut microbiota between the TD and C-ASD groups. The area of the circle indicates the quantity of OTUs. There were 1639 shared OTUs and 1316 unique OTUs in the TD group and 353 unique OTUs in the C-ASD group (Fig. 2D).

Decreased bacterial richness and diversity in the C-ASD group. (A) Comparison of bacterial alpha diversity indexes, including Observe, Chao1, ACE, Shannon, Simpson, and J. (B) ANOSIM was used to analyse the significant differences in colony distribution between the C-ASD group and the TD group (R = 0.0196, p = 1e-04). (C) PCoA revealing the bacterial communities between t the C-ASD group and the TD group on the PCoA1 vs PCoA2 axis (p = 0.001). (D) Venn diagram showing the differences in bacterial community structures between samples from the C-ASD group and the TD group.
Discriminative taxa between the TD and the C-ASD group
LEfSe can be used to analyse the differences between groups and identify differences in microbial species. In the TD group and the C-ASD group, the Linear discriminant analysis threshold was set at 3 to screen the 16 characteristic taxa of the corresponding groups. Ruminococcaceae_UCG_002, Erysipelotrichaceae_UCG_003, Phascolarctobacterium, Megamonas, Ruminiclostridium_5, Parabacteroides, Prevotella_2, Fusobacterium, and Prevotella_9 were enriched in the C-ASD group, and Anaerostipes, Lactobacillus, Ruminococcus_gnavus_group, Lachnospiraceae_NK4A136_group, Ralstonia, Eubacterium_eligens_group, and Ruminococcus_1 were abundant in the TD group (Fig. 3 and 4). The relative abundances of Solibacterales, Corynebacterium, Brevibacterium, etc., were significantly higher in the TD group than in the C-ASD group. Furthermore, the relative abundances of Acidobacteriales, Enorma, Barnesiella, etc., were significantly higher in the C-ASD group than in the TD group (Appendix 3: Figure A2). Furthermore, the 20 distinguished pathways with lower enrichment in the C-ASD group were identified to determine the potential interaction mode (Appendix 4: Figure A3).

Discriminative taxa between the TD group and C-ASD group. LEfSe can be used to analyse the differences between groups and identify different microbial species, and this information can be used to develop biomarkers and promote other studies. In the above two syndrome types, the Linear discriminant analysis threshold was set as 3 to screen the characteristic flora of the corresponding syndrome types.

The relative abundance genus levels (LDA > 3, LEfSe) of gut microbiota between the TD group and the C-ASD group.
Comparison of SCFAs levels detected in faeces between the TD and the C-ASD group
We compared the SCFAs levels in the faeces of the TD group and the C-ASD group, including acetate, PPA, butyrate, valerate, isobutyrate, isovalerate, and hexanoate. The results showed that the faecal PPA levels in the C-ASD group were significantly higher than those in the TD group (p < 0.05), and there was no significant difference in acetate, butyrate, valerate, isobutyrate, isovalerate, and hexanoate levels between TD and C-ASD group (Fig. 5 and Appendix 5: Figure A4).

The levels of acetic, propionate, butyrate, and valerate acid in feces. Each value was presented as the mean ± SEM. p < 0.05.*.
Association among differential bacterial taxa, SCFAs levels and clinical manifestations
The associations among the differential bacterial taxa, SCFAs levels, and clinical manifestations were investigated with Spearman’s correlation analysis (Fig. 6 and Appendix 6: Table A2). Notably, the level of acetate was positively correlated with Parabacteroides (r = 0.340, P = 0.032), Ruminococcaceae_UCG-002 (r = 0.328, P = 0.039), Lachnospiraceae_NK4A136_group (r = 0.314, P = 0.049). PPA, a SCFA elevated in C-ASD group, was negatively associated with TD group enriched genera, such as Lachnospiraceae_NK4A136_group (r = − 0.502, P = 0.001), Anaerostipes (r = − 0.590, P = 6.06E − 05), Ralstonia (r = − 0.392, P = 0.012) , while PPA was positively associated with Prevotella_9 (r = 0.361, P = 0.022) , Phascolarctobacterium (r = 0.367, P = 0.020), Autism Behaviour Scale (ABC) (r = 0.430, P = 0.005) and Children’s Autism Rating Scale (CARS) (r = 0.492, P = 0.001) respectively. In addition, isovalerate was positively associated with ABC (r = 0.335, P = 0.034). Insufficient Lactobacillus (observed in C-ASD group) was negatively correlated with increased PPA (r = − 0.901, P = 2.21E − 15), isobutyrate (r = − 0.317, P = 0.046), ABC (r = − 0.319, P = 0.045) and CARS (r = − 0.353, P = 0.025). Anaerostipes, the genus increased in TD group, was negatively correlated with CARS (r = − 0.416, P = 0.008).

Integrative network among the discriminative genera, short-chain fatty acids and the clinical indexes. Network revealed significant (P < 0.05) between differentially abundant taxa or SCFAs metabolites and clinical indexes in TD (n = 40) and C-ASD (n = 40), respectively. Lines connecting nodes indicate positive (blue) or negative (red) correlations.
To further understand whether these disease-associated taxa and PPA contribute to disease severity, we tested for their correlations with clinical parameters using partial correlation analysis, controlling for age, BMI, birth weight, maternal age and CEBQ scores (including food fussiness, satiety responsiveness, and desire to drink). ABC score was negatively associated with Lactobacillus (r = − 0.382, P = 0.031) (Fig. 7A). However, Lactobacillus has no association of CARS scores by partial correlation analysis (Fig. 7B). The PPA levels, which were higher in the C-ASD group, were negatively correlated with the abundance of Lactobacillus taxa (r = − 0.447, P = 0.010) (Fig. 7C). We also found that PPA was positively correlated with ABC (r = 0.570, P = 0.001) and CARS (r = 0.391, P = 0.027) respectively (Fig. 7D,E), meanwhile isovalerate exhibited a tendency of positive association with ABC (r = 0.360, P = 0.043) (Fig. 7F). In conclusion, we found that PPA was positively correlated with the severity of ASD symptoms.

Relationship between differentially abundant correlative taxa or propionate and ASD scores (ABC and CARS) in C-ASD group (n = 40) by partial correlation analysis.
Classification model
To investigate the potential utility of gut microbial and SCFAs profiles in C-ASD prediction, we built random forest models based on faecal taxonomic or SCFAs features to discriminate them. First, 16 candidate bacteria were selected according to the LEfSe analysis (LDA > 3). The discriminant model based on the 16 representative genera effectively distinguished the C-ASD group from the TD group (AUC = 0.924, 95% CI: 0.896–0.953) through fivefold cross-validation of the random forest model. However, the PPA-derived model did not improve the classification accuracy (AUC = 0.62, 95% CI: 0.565–0.675) (Fig. 8). Comparatively, the integration of PPA and 16 genera features showed similar accuracy in distinguishing C-ASD from TD (AUC = 0.924, 95% CI: 0.895–0.952) (Fig. 8). Taken together, these data indicated that the predictive models based on 16 key discriminatory bacterial taxa performed better than those based on PPA in discriminating C-ASD from TD.

Receiver operating characteristic curve of training data based on selected features.
Discussion
This study investigated associations among the gut microbiota, SCFAs levels and C-ASD in Chinese children. Bacterial richness and diversity were decreased in Chinese children with C-ASD. Nine gut microbiota taxa were enriched in the C-ASD group, and seven gut microbiota taxa were enriched in the TD group. Faecal PPA levels were significantly higher in patients with C-ASD than in the TD group, and PPA levels were negatively correlated with Lactobacillus abundance, which in turn was negatively correlated with the severity of ASD.
Alterations of the gut microbiota in ASD have been described in previous reports. The abundance of Lactobacillus, Clostridia Cluster 1, and Desulfovibrio was increased in children with ASD20. Sutterella, Odoribacter and Butyricimonas were more abundant in the ASD group; nevertheless, the abundance of Veillonella and Streptococcus were observably reduced27. In the genera level, there was a higher abundance of the genera Ruminococcus, Bacteroides, Parabacteroides, and Phascolarctobacterium in individuals with C-ASD28,29. However, in our research, we found that the abundances of Ruminococcaceae_UCG_002, Erysipelotrichaceae_UCG_003, Phascolarctobacterium, Megamonas, Ruminiclostridium_5, Parabacteroides, Prevotella_2, Fusobacterium, and Prevotella_9 were higher in the C-ASD group. This inconsistency may be due to differences in genetic factors, disorder heterogeneity, dynamic microbiota development, locations, dietary habits, lifestyle, age, and detection methods.
The pathways involved in the microbiota-gut-brain axis in ASD include neuronal pathways, neural immune pathways and chemical signalling pathways13. The gut microbiota, such as Lactobacillus rhamnosus JB130 and L. reuteri31, influence the brain via the vagal nerve pathway. Via chemical signalling pathways, the gut microbiota can also influence with the brain through microbial metabolites such as SCFAs, bile acids, and gamma aminobutyric acid13. The brains of mice colonized with ASD-associated microbiota displayed alternative splicing of ASD-relevant genes, and the gut microbiota adjust behaviours in mice through neuroactive metabolites (SCFAs), which contribute to the pathophysiology of ASD via the brain-gut axis32. In this study, there were no significant differences in acetate, butyrate, valerate, isobutyrate, isovalerate, and hexanoate levels between the TD group and the C-ASD group. We found that PPA levels were elevated in the C-ASD group compared with the TD group. In line with another study, the total amount of SCFAs was lower in ASD children, but PPA and acetate were found at higher levels in ASD children. In addition, the abundance of Bacteroides spp. was correlated with PPA levels in ASD individuals33. The association of PPA levels with the gut microbiota in the previous study was consistent with our study, in which PPA levels were negatively correlated with Lactobacillus abundance and positively correlated with Prevotella_9, Phascolarctobacterium, Erysipelatoclostridium, Paraprevotella, Maihella, and Comamonas abundance. In addition, Alcaligenaceae and Porphyromonadaceae abundance was also positively correlated with PPA levels34. In previous studies35, 111 strains of Clostridium perfringens, a PPA-related bacteria, have been isolated from faeces of children with ASD. Meanwhile, it has been demonstrated that Beta2 toxin encoding cpb2 gene is associated with gastrointestinal diseases and significantly more common in the strains of Clostridium perfringens. As evidence indicates that Clostridia spp. produce PPA and worsen autistic symptoms, we inferred that the gut microbiota positively correlated with PPA levels may be potentially harmful microbiota in ASD. The survey on the isolation, identification and characterization of the PA-associated bacteria from the fecal samples of healthy and ASD children based on the traditional plate method was conducted by Yu et al.36. And, a PPA-resistant L. plantarum strain 6–1 was successfully obtained.
Since the level of PPA was significantly higher in the C-ASD group and positively correlated with ABC and CARS scores in the current study, we hypothesized that PPA levels would be associated with the severity of ASD symptoms. Studies have demonstrated that rat models of ASD can be generated using various routes of PPA administration37,38,39. PPA-induced behavioural effects were demonstrated to coincide with ASD symptoms in patients. When PPA solution was injected into the ventricles of rat brains, the rats showed autism-like behaviour, and the structure of their nerve cells was altered38,39. PPA plays a significant role in ASD causation by altering several developmental molecular pathways, such as the PTEN/Akt, MAPK/ERK, mTOR/Gskβ, and cytokine-activated pathways, at the prenatal and neonatal stages40,41. A study showed that excessive PPA levels directly damaged neurons and was linked to neurological disorders42. From a physiological perspective, PPA can pass through the blood‒brain barrier and influence the central nervous system39. These results indicated that in C-ASD children, excessive PPA levels (as measured in faeces) might be an important factor aggravating ASD symptoms.
Regarding eating patterns, although no significant difference was found between the two groups in CEBQ scores, the C-ASD group showed more food fussiness, satiety responsiveness and desire to drink. A growing number of studies have established links between GI disorders and the gut microbiota in children with ASD. As a probiotic, Lactobacillus spp. can regulate the balance of intestinal microorganisms and help relieve diarrhoea and constipation43. Lactobacillus spp. also reduce the symptoms of constipation by shortening intestinal transit time, increasing the abundance of beneficial bacteria, and increasing faecal moisture44,45,46,47. Moreover, both animal and clinical studies have shown that probiotics can change the gut microbiota and thereby affect central nervous system function through the gut-brain axis. As a nondrug and relatively risk-free option, probiotics have gradually become an adjuvant treatment for children with ASD48,49,50. A study showed that after patients ate or were injected with a solution that contained Lactobacillus spp., the abundance of the gut microbiota changed significantly: the abundance of Firmicutes taxa, which produce PPA, was decreased, and the diversity of the gut microbiota was increased51. Lactobacillus spp. were shown to alleviate anxiety-like behaviours and increase dopamine levels in the prefrontal cortex in mice exposed to early life stress52. Probiotic treatment such as Bifidobacteria and Lactobacilli reduces the autistic-like excitation/inhibition imbalance in juvenile hamsters induced by orally administered propionic acid and clindamycin, mostly through increasing depleted γ-aminobutyric acid and Mg2++ and decreasing the excitatory neurotransmitter, glutamate53. Similarly, Aabed et al.54 found that probiotic/prebiotic treatments showed ameliorative effects on clindamycin and propionic acid-induced oxidative stress and altered gut microbiota in a rodent model of autism; however, lactobacillus had the strongest effect. In a four-week, randomized, double-blind, placebo-controlled study55, Lactobacillus spp. have also been shown to reduce the symptoms of ASD. Additionally, it has been shown that patients with ASD who take probiotics (including Lactobacillus spp.) have lower concentrations of PPA in the faeces56. These studies hint at the potential to administer probiotic interventions to reduce GI and ASD symptoms57. Therefore, a promising approach to enhance treatment efficacy in pediatric patients with C-ASD in the southern expanse of China may involve the application of a strategy utilizing PPA-resistant Lactobacillus.
Several scales, such as the CARS and ABC, are available to assess behaviours and symptoms associated with ASD. However, diagnosing ASD has always been difficult, and the condition is recognized at a late stage for treatment to have a major effect. Several studies have focused on biomarkers for determining ASD risk or assisting with diagnosis, such as proteomic analysis58, epigenome-wide analysis59, single-nucleotide polymorphism60, maackia amurensis lectin-II binding glycoproteins61, and the gut microbiota62,63,64,65. In our study, the use of microbial markers for the prediction of C-ASD was comprehensively assessed by 16 specific genera and PPA. The best-performing model achieved a high accuracy (AUC = 0.924) and included 16 important features that distinguished C-ASD from TD. Our results imply that the gut microbiome might provide new biomarkers to predict C-ASD.
There are some limitations of our study. First, we did not control for confounding factors of geographical differences, which can affect the gut microbiome. All included subjects were recruited from a city along the southeastern coast of China. Thus, the topography, climatic characteristics, and dietary patterns are similar. Second, intestinal bacteria were analysed based on 16S rRNA rather than metagenomics. Third, given the small sample size, more studies are needed to confirm these findings, as these data did not permit subgroup analysis. Since ASD is a highly heterogeneous disorder, it is more reasonable to perform a subgroup analysis to assess factors such as symptom severity. Finally, due to a cross-sectional study, the causal relationship among the gut microbiota, SCFAs levels and C-ASD remains unknown, but a complex association was observed.
Conclusion
In summary, we identified different profiles of the gut microbiota and SCFAs in C-ASD and TD children. We investigated the associations among C-ASD clinical parameters, differential bacteria and PPA levels. These differences provide strong evidence that specific genera Lactobacillus within the gut represent a link between intestinal bacteria and C-ASD in children and could provide a theoretical basis for the treatment of C-ASD in the future.
Materials and methods
Human participants
Ethical approval was granted by the Ethics Committee of Zhongshan Hospital of Xiamen University (xmzsyyky (2021–171)) and registered in the Chinese Clinical Trial Registry (www.chictr.org.cn; trial registration number ChiCTR2100052106). All experiments were performed in accordance with relevant guidelines and regulations. Participants were recruited from Xiamen, Fujian Province, China, between November 1, 2021 and November 1, 2022. Inclusion criteria of the C-ASD group were as follows: (1) aged 3 to 10 years; (2) diagnosed with ASD according to the Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-V) criteria (CARS score ≥ 30 and ABC total score ≥ 67); and the International Statistical Classification of Diseases and Related Health Problems (ICD-10). (3) presence of GI comorbidities according to the Childhood Functional Gastrointestinal Disorders criteria (2016)66. (4) The guardians of each patient signed an informed consent form to allow the investigator to collect clinical data, faeces, and other samples. Two experienced child neuropsychiatrists determined the diagnosis of ASD. All parents were asked to complete the following questionnaires: CARS, ABC and CEBQ. Sex- and age-matched TD control children without GI disorders were included in the trial. The inclusion criteria for the TD children were as follows: (1) aged 3 to 10 years; and (2) in good health, without a clearly diagnosed disease. We adopted the following exclusion criteria before faecal sample collection: (1) definite diagnosis of Rett syndrome; (2) a clear history of brain injuries, cerebral palsy, encephalitis or other organic brain diseases; (3) a family history of serious mental illness or substance abuse; (4) use of probiotics, antibiotics, acid suppressors or other drugs affecting the gut microbiota in the last three months; (5) the presence of intestinal organic diseases, such as congenital megacolon, intestinal obstruction, intussusception and developmental disorder-related genetic diseases; (6) developmental quotient score < 35; or (7) participation in other clinical trials that would affect the evaluation of the study results. The confidentiality of participants results was maintained by de-identifying all samples. The participant's name was not used in all sections of the manuscript.
Determination of clinical parameters
We collected basic information from all subjects (age, birth weight, height, and weight) and calculated the BMI using height and weight. We also interviewed the parents of the children to collect information on maternal age, gestational age, delivery mode, feeding patterns, maternal educational level, maternal smoking history, maternal drinking history, paternal educational level, paternal smoking history, and paternal drinking history. Children in the C-ASD group were assessed with the CARS and ABC. All children were evaluated with CEBQ.
Sequencing and bioinformatics
Faecal samples were collected in sterile plastic cups and stored at − 80 °C for 1 h until further processing67. Faecal microbial DNA was extracted using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). PCR amplification was conducted using an ABI 2720 Thermal Cycler (Thermo Fisher Scientific, USA). We used Multiskan™ GO spectrophotometry (Thermo Fisher Scientific, USA) to quantify bacterial genomic DNA as the template for amplification of the V3-V4 hypervariable region of the 16S rRNA gene in three replicate reactions with forwards (Illumina adapter sequence 5’-CCTACGGGNBGCASCAG-3’) and reverse (Illumina adapter sequence 5’-GGACTACNVGGGTWTCTAAT-3’) primers. Replicate PCR products were pooled and purified with Agencourt AMPure XP magnetic beads (Beckman Coulter, USA). A TopTaq DNA Polymerase kit (TraTransgenichina) was used. The purity and concentration of sample DNA were assessed using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, USA). Paired-end sequencing was performed by Treatgut Biotechnology Co., Ltd. with a HiSeq 2500 (Illumina, San Diego, CA, USA) with PE 250 bp reagents. After sequencing, raw paired-end reads were assembled using FLASH68 with the default parameters. Primers were removed using cutadapt, and clean tags were obtained by removing the lower reads using cutadapt69. To assign de novo operational taxonomic units (OTUs), we removed chimeric sequences and clustered sequences with 97% similarity using Research (V10.0.240)70. The representative sequences of OTUs were aligned to the SILVA132 database for taxonomic classification by RDP Classifier71 and aggregated to various taxonomic levels.
SCFAs measurement
Frozen faecal samples were mixed with 1 mL of ice-cold saline, homogenized in a ball mill (4 min, 40 Hz), and then ultrasonically processed for 5 min in ice water, a process which was repeated three times. The supernatant was obtained by centrifugation (5,000 rpm, 20 min, 4 °C) and added to extracting solution containing 2-methylpentanoic acid (25 mg/L in methyl tert-butyl ether) as an internal standard. The mixture was then fully mixed by vortexing and oscillation and centrifuged for 15 min (10,000 rpm, 4 °C). After centrifugation, the supernatant was transferred into a fresh vial for gas chromatography‒mass spectrometry analysis. Gas chromatography‒mass spectrometry analysis was performed using an Agilent gas chromatograph coupled with an Agilent 5977B mass spectrometer. Helium was used as the carrier gas, and the initial temperature was set to 80 °C (1 min), subsequently increased to 200 °C (in 5 min), and then maintained (1 min) at 240 °C. The injection and ion source temperatures were 240 and 230 °C, respectively. The electron impact energy was -70 Eva. The mass spectrometry data were acquired in Scan/SIM mode.
Statistical analyses and visualization
The rarefaction curves constructed from the sequenced data were basically stable, indicating that the sequenced data were basically stable at this sequencing depth (Appendix 1: Figure A1). The alpha diversity indices (Shannon, Simpson, ACE, and Chao1 indices) and bacterial richness (observed OTUs) and evenness (J) were calculated based on the OTU tables of the study. Detection of significant differences between the TD and C-ASD groups was performed with the Wilcoxon test. Differences in community structure across samples (beta diversity) were visualized by principal coordinates analysis (PCoA) plots based on Bray‒Curtis distance. Significance tests were determined using permutational multivariate analysis of variance (PERMANOVA) with 999 permutations in vegan72. Linear discriminant analysis effect size (LEfSe)73 was performed to identify taxa with differential abundance between the TD and C-ASD groups. We further explored the correlations among clinical data, abundance of different genera (LEfSe, LDA > 3) and faecal SCFAs by Spearman’s correlation analyses. Partial correlation analysis (PResiduals package) was used to evaluate associations between differentially abundant correlative taxa (P < 0.05, Spearman’s correlation analyses) or SCFAs and ASD scores (ABC and CARS) in C-ASD group. To evaluate functional differences in the gut microbiomes of the TD versus the C-ASD groups, we used PICRUSt74 to calculate the microbial abundances, assigned metabolic pathways to the gut microbiomes using KEGG and COG analyses, and then tested the differences between the two groups. To train multivariable statistical models for the prediction of TD and C-ASD groups, two levels of bacterial and SCFAs features were combined to develop prediction models using the random Forest R package v4.6–14. Finally, all predictions were used to calculate the area under the receiver operating characteristics curve (AUC) using the pROC R package v1.17.01. All statistical and correlational analyses were conducted in R (v3.6.0)75. Figures were plotted mainly using ggplot2 (v3.0.0)76.
Ethical approval
This study was approved by the Ethics Committee of Zhongshan Hospital of Xiamen University (xmzsyyky (2021–171)) and registered in the Chinese Clinical Trial Registry (www.chictr.org.cn; trial registration number ChiCTR2100052106). All participants and/or their parents provided written informed consent for their participation in the study.
Data availability
The datasets generated during and analysed during the current study are available from the corresponding author upon reasonable request. All sequence data is available from NCBIs Sequence Read Archive (SRA), Bio Project ID: PRJNA932561.
References
-
Hyman, S. L., Levy, S. E. & Myers, S. M. Identification, evaluation, and management of children with autism spectrum disorder. Pediatrics 145, 3447. https://doi.org/10.1542/peds.2019-3447 (2020).
Google Scholar
-
Maenner, M. J. et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years – autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill. Summ. 70, 7011. https://doi.org/10.15585/mmwr.ss7011a1 (2021).
Google Scholar
-
Wang, F. et al. The prevalence of autism spectrum disorders in China: A comprehensive meta-analysis. Int. J. Biol. Sci. 14, 717–725. https://doi.org/10.7150/ijbs.24063 (2018).
Google Scholar
-
Gorrindo, P. et al. Gastrointestinal dysfunction in autism: Parental report, clinical evaluation, and associated factors. Autism Res. 5, 101–108. https://doi.org/10.1002/aur.237 (2012).
Google Scholar
-
Lefter, R., Ciobica, A., Timofte, D., Stanciu, C. & Trifan, A. A descriptive review on the prevalence of gastrointestinal disturbances and their multiple associations in autism spectrum disorder. Medicina 56, 11 (2020).
Google Scholar
-
McElhanon, B. O., McCracken, C., Karpen, S. & Sharp, W. G. Gastrointestinal symptoms in autism spectrum disorder: A meta-analysis. Pediatrics 133, 872–883. https://doi.org/10.1542/peds.2013-3995 (2014).
Google Scholar
-
Holingue, C., Newill, C., Lee, L. C., Pasricha, P. J. & Daniele Fallin, M. Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism Res. 11, 24–36. https://doi.org/10.1002/aur.1854 (2018).
Google Scholar
-
Dovgan, K., Gynegrowski, K. & Ferguson, B. J. Bidirectional relationship between internalizing symptoms and gastrointestinal problems in youth with autism spectrum disorder. J. Autism Dev. Disord. https://doi.org/10.1007/s10803-022-05539-6 (2022).
Google Scholar
-
Kang, D. W. et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 5, 10. https://doi.org/10.1186/s40168-016-0225-7 (2017).
Google Scholar
-
Saurman, V., Margolis, K. G. & Luna, R. A. Autism spectrum disorder as a brain-gut-microbiome axis disorder. Dig. Dis. Sci. 65, 818–828. https://doi.org/10.1007/s10620-020-06133-5 (2020).
Google Scholar
-
Srikantha, P. & Mohajeri, M. H. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int. J. Mol. Sci. 20, 2115. https://doi.org/10.3390/ijms20092115 (2019).
Google Scholar
-
Wong, G. C., Montgomery, J. M. & Taylor, M. W. In Autism Spectrum Disorders (ed A. M. Grabrucker) (Exon Publications, 2021).
-
Yu, Y. & Zhao, F. Microbiota-gut-brain axis in autism spectrum disorder. J. Genet. Genomics 48, 755–762. https://doi.org/10.1016/j.jgg.2021.07.001 (2021).
Google Scholar
-
Kushak, R. I. et al. Evaluation of intestinal function in children with autism and gastrointestinal symptoms. J. Pediatr. Gastroenterol. Nutr. 62, 687–691. https://doi.org/10.1097/mpg.0000000000001174 (2016).
Google Scholar
-
Bermudez-Martin, P. et al. The microbial metabolite p-Cresol induces autistic-like behaviors in mice by remodeling the gut microbiota. Microbiome 9, 157. https://doi.org/10.1186/s40168-021-01103-z (2021).
Google Scholar
-
Kang, D. W. et al. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe 49, 121–131. https://doi.org/10.1016/j.anaerobe.2017.12.007 (2018).
Google Scholar
-
Wang, M. et al. Alterations in gut glutamate metabolism associated with changes in gut microbiota composition in children with autism spectrum disorder. mSystems 4, 18. https://doi.org/10.1128/mSystems.00321-18 (2019).
Google Scholar
-
Liu, F. et al. Altered composition and function of intestinal microbiota in autism spectrum disorders: A systematic review. Transl. Psychiat. 9, 43. https://doi.org/10.1038/s41398-019-0389-6 (2019).
Google Scholar
-
Strati, F. et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 5, 24. https://doi.org/10.1186/s40168-017-0242-1 (2017).
Google Scholar
-
Tomova, A. et al. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 138, 179–187. https://doi.org/10.1016/j.physbeh.2014.10.033 (2015).
Google Scholar
-
Kang, D. W. et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One 8, e68322. https://doi.org/10.1371/journal.pone.0068322 (2013).
Google Scholar
-
Liu, S. et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 9, 287. https://doi.org/10.1038/s41598-018-36430-z (2019).
Google Scholar
-
Wang, H., Liu, S., Xie, L. & Wang, J. Gut microbiota signature in children with autism spectrum disorder who suffered from chronic gastrointestinal symptoms. BMC Pediatr. 23, 476. https://doi.org/10.1186/s12887-023-04292-8 (2023).
Google Scholar
-
Zhao, Y. et al. Altered gut microbiota as potential biomarkers for autism spectrum disorder in early childhood. Neuroscience 523, 118–131. https://doi.org/10.1016/j.neuroscience.2023.04.029 (2023).
Google Scholar
-
Wong, O. W. H. et al. Disentangling the relationship of gut microbiota, functional gastrointestinal disorders and autism: A case–control study on prepubertal Chinese boys. Sci. Rep. 12, 10659. https://doi.org/10.1038/s41598-022-14785-8 (2022).
Google Scholar
-
Alshammari, M. K., AlKhulaifi, M. M., Al Farraj, D. A., Somily, A. M. & Albarrag, A. M. Incidence of Clostridium perfringens and its toxin genes in the gut of children with autism spectrum disorder. Anaerobe 61, 102114. https://doi.org/10.1016/j.anaerobe.2019.102114 (2020).
Google Scholar
-
Zhang, M., Ma, W., Zhang, J., He, Y. & Wang, J. Analysis of gut microbiota profiles and microbe-disease associations in children with autism spectrum disorders in China. Sci. Rep. 8, 13981. https://doi.org/10.1038/s41598-018-32219-2 (2018).
Google Scholar
-
Dan, Z. et al. Altered gut microbial profile is associated with abnormal metabolism activity of autism spectrum disorder. Gut. Microbes 11, 1246–1267. https://doi.org/10.1080/19490976.2020.1747329 (2020).
Google Scholar
-
Iglesias-Vázquez, L., Van Ginkel Riba, G., Arija, V. & Canals, J. Composition of gut microbiota in children with autism spectrum disorder: A systematic review and meta-analysis. Nutrients 12, 30978. https://doi.org/10.3390/nu12030792 (2020).
Google Scholar
-
Bravo, J. A. et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U. S. A. 108, 16050–16055. https://doi.org/10.1073/pnas.1102999108 (2011).
Google Scholar
-
Sgritta, M. et al. Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246–259. https://doi.org/10.1016/j.neuron.2018.11.018 (2019).
Google Scholar
-
Sharon, G. et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 177, 1600-1618.e1617. https://doi.org/10.1016/j.cell.2019.05.004 (2019).
Google Scholar
-
De Angelis, M. et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PloS one 8, e76993. https://doi.org/10.1371/journal.pone.0076993 (2013).
Google Scholar
-
Borghi, E. et al. Rett syndrome: A focus on gut microbiota. Int. J. Mol. Sci. 18, 64122 (2017).
Google Scholar
-
Góra, B. et al. Toxin profile of fecal Clostridium perfringens strains isolated from children with autism spectrum disorders. Anaerobe 51, 73–77. https://doi.org/10.1016/j.anaerobe.2018.03.005 (2018).
Google Scholar
-
Yu, R. et al. Isolation, identification and characterization of propionic acid bacteria associated with autistic spectrum disorder. Microbial. Pathog. 147, 104371. https://doi.org/10.1016/j.micpath.2020.104371 (2020).
Google Scholar
-
MacFabe, D. F. et al. Neurobiological effects of intraventricular propionic acid in rats: Possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav. Brain Res. 176, 149–169. https://doi.org/10.1016/j.bbr.2006.07.025 (2007).
Google Scholar
-
Thomas, R. H. et al. The enteric bacterial metabolite propionic acid alters brain and plasma phospholipid molecular species: further development of a rodent model of autism spectrum disorders. J. Neuroinflammation 9, 153. https://doi.org/10.1186/1742-2094-9-153 (2012).
Google Scholar
-
Choi, J. et al. Pathophysiological and neurobehavioral characteristics of a propionic acid-mediated autism-like rat model. PLoS One 13, e0192925. https://doi.org/10.1371/journal.pone.0192925 (2018).
Google Scholar
-
Sharma, A. R. et al. Valproic acid and propionic acid modulated mechanical pathways associated with autism spectrum disorder at prenatal and neonatal exposure. CNS Neurol. Disord. Drug Targets 21, 399–408. https://doi.org/10.2174/1871527320666210806165430 (2022).
Google Scholar
-
Choi, H., Kim, I. S. & Mun, J. Y. Propionic acid induces dendritic spine loss by MAPK/ERK signaling and dysregulation of autophagic flux. Mol. Brain 13, 86. https://doi.org/10.1186/s13041-020-00626-0 (2020).
Google Scholar
-
Al-Lahham, S. H., Peppelenbosch, M. P., Roelofsen, H., Vonk, R. J. & Venema, K. Biological effects of propionic acid in humans; Metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta 1175–1183, 2010. https://doi.org/10.1016/j.bbalip.2010.07.007 (1801).
Google Scholar
-
Saviano, A. et al. Lactobacillus Reuteri DSM 17938 (Limosilactobacillus reuteri) in diarrhea and constipation: Two sides of the same coin?. Med. Kaunas 57, 57070422. https://doi.org/10.3390/medicina57070643 (2021).
Google Scholar
-
Tang, T. et al. Bifidobacterium lactis TY-S01 prevents loperamide-induced constipation by modulating gut microbiota and its metabolites in mice. Front. Nutr. 9, 890314. https://doi.org/10.3389/fnut.2022.890314 (2022).
Google Scholar
-
Dimidi, E., Christodoulides, S., Fragkos, K. C., Scott, S. M. & Whelan, K. The effect of probiotics on functional constipation in adults: A systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 100, 1075–1084. https://doi.org/10.3945/ajcn.114.089151 (2014).
Google Scholar
-
Agrawal, A. et al. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN-173 010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment. Pharmacol. Therap. 29, 104–114. https://doi.org/10.1111/j.1365-2036.2008.03853.x (2009).
Google Scholar
-
Yang, Z. et al. Dietary synbiotic ameliorates constipation through the modulation of gut microbiota and its metabolic function. Food Res. Int. 147, 110569. https://doi.org/10.1016/j.foodres.2021.110569 (2021).
Google Scholar
-
Partty, A., Kalliomaki, M., Wacklin, P., Salminen, S. & Isolauri, E. A possible link between early probiotic intervention and the risk of neuropsychiatric disorders later in childhood: A randomized trial. Pediatr. Res. 77, 823–828. https://doi.org/10.1038/pr.2015.51 (2015).
Google Scholar
-
Lammert, C. R. et al. Cutting edge: Critical roles for microbiota-mediated regulation of the immune system in a prenatal immune activation model of autism. J. Immunol. Baltim. Md 1950 201, 845–850. https://doi.org/10.4049/jimmunol.1701755 (2018).
Google Scholar
-
Desbonnet, L. et al. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 170, 1179–1188. https://doi.org/10.1016/j.neuroscience.2010.08.005 (2010).
Google Scholar
-
Li, X. R. et al. Gut microbiota alterations from three-strain yogurt formulation treatments in slow-transit constipation. Can. J. Infect. Dis. Med. Microbiol. 2020, 4583973. https://doi.org/10.1155/2020/4583973 (2020).
Google Scholar
-
Liu, Y.-W. et al. Psychotropic effects of Lactobacillus plantarum PS128 in early life-stressed and naïve adult mice. Brain Res. 1631, 1–12. https://doi.org/10.1016/j.brainres.2015.11.018 (2016).
Google Scholar
-
El-Ansary, A. et al. Probiotic treatment reduces the autistic-like excitation/inhibition imbalance in juvenile hamsters induced by orally administered propionic acid and clindamycin. Metab. Brain Dis. 33, 1155–1164. https://doi.org/10.1007/s11011-018-0212-8 (2018).
Google Scholar
-
Aabed, K. et al. Ameliorative effect of probiotics (Lactobacillus paracaseii and Protexin®) and prebiotics (propolis and bee pollen) on clindamycin and propionic acid-induced oxidative stress and altered gut microbiota in a rodent model of autism. Cell. Mol. Biol. Noisy Grand France 65, 1–7 (2019).
Google Scholar
-
Liu, Y. W. et al. Effects of Lactobacillus plantarum PS128 on children with autism spectrum disorder in Taiwan: A randomized, double-blind. Placebo Control. Trial. Nutr. 11, 4080255. https://doi.org/10.3390/nu11040820 (2019).
Google Scholar
-
Adams, J. B., Johansen, L. J., Powell, L. D., Quig, D. & Rubin, R. A. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 11, 22. https://doi.org/10.1186/1471-230x-11-22 (2011).
Google Scholar
-
Karhu, E. et al. Nutritional interventions for autism spectrum disorder. Nutr. Rev. 78, 515–531. https://doi.org/10.1093/nutrit/nuz092 (2020).
Google Scholar
-
Hewitson, L. et al. Blood biomarker discovery for autism spectrum disorder: A proteomic analysis. PloS one 16, e0246581. https://doi.org/10.1371/journal.pone.0246581 (2021).
Google Scholar
-
Bahado-Singh, R. O. et al. Artificial intelligence analysis of newborn leucocyte epigenomic markers for the prediction of autism. Brain Res. 1724, 146457. https://doi.org/10.1016/j.brainres.2019.146457 (2019).
Google Scholar
-
Ghafouri-Fard, S. et al. Application of single-nucleotide polymorphisms in the diagnosis of autism spectrum disorders: a preliminary study with artificial neural networks. J. Mol. Neurosci. MN 68, 515–521. https://doi.org/10.1007/s12031-019-01311-1 (2019).
Google Scholar
-
Qin, Y. et al. Serum glycopattern and Maackia amurensis lectin-II binding glycoproteins in autism spectrum disorder. Sci. Rep. 7, 46041. https://doi.org/10.1038/srep46041 (2017).
Google Scholar
-
Wan, Y. et al. Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut 71, 910–918. https://doi.org/10.1136/gutjnl-2020-324015 (2022).
Google Scholar
-
Lou, M. et al. Deviated and early unsustainable stunted development of gut microbiota in children with autism spectrum disorder. Gut 71, 1588–1599. https://doi.org/10.1136/gutjnl-2021-325115 (2022).
Google Scholar
-
Ding, X. et al. Gut microbiota changes in patients with autism spectrum disorders. J. Psychiatr. Res. 129, 149–159. https://doi.org/10.1016/j.jpsychires.2020.06.032 (2020).
Google Scholar
-
Xu, Y. et al. Leveraging existing 16SrRNA microbial data to define a composite biomarker for autism spectrum disorder. Microbiol. Spectr. 10, e0033122. https://doi.org/10.1128/spectrum.00331-22 (2022).
Google Scholar
-
Hyams, J. S. et al. Childhood functional gastrointestinal disorders: Child/adolescent. Gastroenterology 150, 1456-1468.e1452. https://doi.org/10.1053/j.gastro.2016.02.015 (2016).
Google Scholar
-
Yang, L. et al. Preservation of the fecal samples at ambient temperature for microbiota analysis with a cost-effective and reliable stabilizer EffcGut. Sci. Total Environ. 741, 140423. https://doi.org/10.1016/j.scitotenv.2020.140423 (2020).
Google Scholar
-
Magoč, T. & Salzberg, S. L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. https://doi.org/10.1093/bioinformatics/btr507%JBioinformatics (2011).
Google Scholar
-
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 200. https://doi.org/10.14806/ej.17.1.200 (2011).
Google Scholar
-
Edgar, R. C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. https://doi.org/10.1038/nmeth.2604 (2013).
Google Scholar
-
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. https://doi.org/10.1128/AEM.00062-07 (2007).
Google Scholar
-
Oksanen, J. et al. Vegan: Community Ecology Package. R package version 2.3–5. (2016).
-
Paulson, J. N., Stine, O. C., Bravo, H. C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. https://doi.org/10.1038/nmeth.2658 (2013).
Google Scholar
-
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. https://doi.org/10.1038/nbt.2676 (2013).
Google Scholar
-
Core, R., Rdct, R., Team, R. & Team, R. J. C. A language and environment for statistical computing. 1, 12–21 (2015).
-
Wickham, H., Chang, W. & Abstracts, R. J. B. ggplot2: Create elegant data visualisations using the grammar of graphics. Adv. Pract. Psycol. Sci. 6(3), 235654566 (2016).
Acknowledgements
We thank all the participants and investigators for their continued dedication to this study. We also thank all members of Treat gut Biotechnology Co., Ltd. (Xiamen, China) and Biotree Biotech Co., Ltd. (Shanghai, China) for participating in 16S sRNA Sequencing, SCFAs measurement and bioinformatics analysis. We would like to thank AJE (www.aje.com) for English language editing.
Funding
This study was found by the National Natural Science Foundation of China (82004433), the Science Foundation of Fujian Province in China (2021J011327).
Author information
Authors and Affiliations
Contributions
Conception and design of the study: J.Q.H, G.H.Z, S.J.W and Y.Y.Z. Collection of data: J.Q.H, L.L, Y.Z.X, W.X.G and X.J.L. Analysis and interpretation of data: M.C., B.Z.Z and R.M.X. Drafting of the manuscript: J.Q.H., X.H.G., and B.H. Critical revision of the manuscript for important intellectual content: G.H.Z., S.W.J., and Y.Y.Z. Administrative support and study supervision: Y.Y.Z. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Information 1.
Supplementary Information 2.
Supplementary Information 3.
Supplementary Information 4.
Supplementary Information 5.
Supplementary Information 6.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
He, J., Gong, X., Hu, B. et al. Altered Gut Microbiota and Short-chain Fatty Acids in Chinese Children with Constipated Autism Spectrum Disorder.
Sci Rep 13, 19103 (2023). https://doi.org/10.1038/s41598-023-46566-2
-
Received: 15 May 2023
-
Accepted: 02 November 2023
-
Published: 04 November 2023
-
DOI: https://doi.org/10.1038/s41598-023-46566-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.