Infection
Characterization of respiratory bacterial co-infection and assessment of empirical antibiotic treatment in patients with COVID-19 at hospital admission
Abstract
Accurate characterization of respiratory bacterial co-infection is critical for guiding empirical antibiotic treatment for hospitalised patients with coronavirus disease 2019 (COVID-19). We retrospectively assessed the clinical and analytical predictors of respiratory bacterial co-infection and described the empirical use of antibiotics in COVID-19 hospitalised patients. Respiratory bacterial co-infection was documented in 6.9% (80/1157) of the patients. The predominant bacteria isolates were Haemophilus influenzae, followed by Streptococcus pneumoniae and Pseudomonas aeruginosa. Respiratory bacterial co-infection was associated with having had a positive culture for a respiratory pathogen in the last year (OR = 25.89), dyslipidaemia (OR = 2.52), heart failure (OR = 7.68), ferritin levels 8.7 × 109/L (OR = 2.4), and patients with chronic obstructive pulmonary disease treated with inhaled corticosteroids (OR = 12.94). Empirical antibiotic treatment was administered in 42.33% of patients, although it declined across the distinct study periods (p < 0.001). Patients admitted to intensive care units harbouring co-infection exhibited worse outcomes and more bacterial secondary infections. In conclusion, respiratory bacterial co-infection prevalence was low, although it could lead to unfavourable outcomes. Moreover, the percentage of empirical antibiotic treatment remained high. The study's findings allowed the identification of several predictors for respiratory bacterial co-infection and could help implement adequate antibiotic stewardship measures.
Introduction
The identification of bacterial co-infection in hospitalised patients with coronavirus disease 2019 (COVID-19) has posed a challenge since the pandemic’s inception. Early-stage studies and meta-analyses have consistently documented low bacterial co-infection rates upon patient admission1,2,3, although some studies have associated bacterial co-infection with adverse outcomes -such as heightened mortality, the need for mechanical ventilation, and prolonged hospital stays-3, 4. However, data on predisposing risk factors for bacterial co-infection remain scarce.
Recent meta-analysis data revealed antibiotic usage prevalence exceeding 70.0% among COVID-19 patients2. In the first months of the pandemic, there was extensive empirical use of azithromycin because its antiviral effect and immunomodulatory properties5. Maintaining vigilance over bacterial co-infection dynamics is essential for steering appropriate empirical antibiotic treatments in hospitalised COVID-19 patients.
To accomplish this, we conducted an analysis on COVID-19 patients admitted at a tertiary hospital between March 2020 and April 2021 to assess the clinical and analytical predictors of respiratory bacterial co-infection. We have also described the empirical antibiotic treatment admission during different periods of the COVID-19 pandemic and evaluated the impact of antibiotic use in patients admitted to intensive care units (ICUs).
Methods
Study design
We performed an observational retrospective study at the Germans Trias i Pujol University Hospital (Badalona, Spain), the designated referral hospital of an urban area with 1,400,000 inhabitants. We have included all patients ≥ 18 years old with a diagnosis of COVID-19 (positive nucleic acid amplification test (NAAT) for SARS-CoV-2 on a nasopharyngeal swab or respiratory tract secretions), and admitted for > 48 h between March 2020 and April 2021. To be eligible for the study, patients should have a positive SARS-CoV-2 NAAT result within three days preceding or two days subsequent admission.
For the analysis of predictors related to respiratory bacterial co-infection, patients were included if they had undergone collection of at least one urine sample for Legionella pneumophila and Streptococcus pneumoniae antigens (Sofia-Quidel, San Diego, USA) within the initial 48 h following admission. Patients were excluded from the analysis if any indications of bacterial co-infection originating from sources other than respiratory infection were detected (refer to the Definitions section).
Data collection
Demographic information, medical history, comorbid conditions, laboratory test, microbiological data, treatment, ICU admission, and outcomes were obtained electronically from an anonymised database known as CORE-COVID, implemented at the hospital. This comprehensive repository aggregates data from eight hospitals and primary healthcare systems, encompassing patients admitted with confirmed COVID-19 diagnoses.
Microbiological procedures and definitions
We performed standard microbiological procedures for the investigation of bacterial and fungal pathogens. S. pneumoniae, L. pneumophila, Haemophilus influenzae, Moraxella catarrhalis, Staphylococcus aureus, Pseudomonas aeruginosa, Achromobacter xylosoxidans, Stenotrophomas maltophila, Burkholderia spp, Aspergillus spp, asa well as microorganisms within the Enterobacterales order and Mycobacteriaceae family were considered as respiratory pathogens.
Respiratory bacterial co-infection was defined as the growth of respiratory pathogens obtained from blood, pleural fluids, good-quality sputum and endotracheal aspirates (> 25 polymorphonuclear leukocytes and < 25 epithelial cells), bronchoalveolar lavages and positive urinary antigen test for S. pneumoniae or L. pneumophila (Sofia-Quidel, San Diego, USA) within the initial 48 h following admission. As mentioned above, patients demonstrating evidence of bacterial or fungal co-infection from sources other than respiratory infection were excluded from the study.
We characterized secondary infections as any positive urine, respiratory tract, or blood cultures emerging 48 h post-admission. We also recorded the proportion of secondary infections attributable to extended-spectrum beta lactamase (ESBL) microorganisms, methicillin-resistant S. aureus (MRSA), vancomycin-resistant enterococci (VRE), and extensively drug-resistant P. aeruginosa were recorded6. These three categories were considered multidrug-resistant infections. Additionally, we documented secondary infections involving Clostridioides difficile. Days of Therapy (DOT) was calculated for each patient and standardised for 100 patient days.
Statistical analysis
First, we compared patients with and without respiratory bacterial co-infection and performed a univariate and multivariate logistic regression model to explore the predictors associated with respiratory bacterial co-infections. Variables with a significance level of < 0.10 in the univariate analysis were included in the multivariate analysis. Odds ratios (ORs) with their 95% CI values were calculated.
Second, we performed a descriptive analysis of empirical antibiotic in hospitalised COVID-19 patients, categorizing them into four distinct study periods aligned with Spain’s first four COVID-19 waves (1st: from March 2020 to May 2020; 2nd: from June 2020 to December 9, 2020; 3rd: from December 10 to March 15, 2021, and 4th: March 16, 2021, to May 1, 2021).
Third, we compared patients admitted to ICUs with and without antibiotic therapy within 48 h of admission to evaluate the impact of empirical antibiotic use within a homogeneous population.
We determined continuous and categorical variables as median (interquartile range (IQR)) and absolute number (percentage), respectively. The Mann–Whitney U, chi-square, and Fisher´s exact tests were performed for descriptive analysis. Significance was set at p < 0.05. We performed the statistical studies using the software SPSS Statistics 24.0 (IBM, Armonk, NY, USA).
Ethics approval
The study was conducted according to the principles of the Declaration of Helsinki and was approved by the Ethics Committee of the Germans Trias i Pujol University Hospital (approval application number PI-21-073). The Ethics Committee of the Germans Trias i Pujol University Hospital waived the requirement for informed consent because of the study’s retrospective nature, and all data were analysed anonymously. All methods of the study were carried out in accordance with institutional guidelines and regulations.
Results
Participant characteristics and respiratory bacterial co-infections
Of the 4858 people in the CORE-COVID database, 2121 (43.66%) were eligible for the study. Within this subset, 1157 (54.55%) underwent assessment for predictors of respiratory bacterial co-infection. Respiratory bacterial co-infection was documented in 6.9% (80/1157) of the patients (6.2% in non-ICU patients and 8.9% in ICU patients; p = 0.001). 3.5% of the patients (41/1157) had a positive urinary antigen test for S. pneumoniae, while one yielded a positive urinary test for L. pneumophila. Furthermore, 3.8% (44/1157) presented a positive bacterial culture from respiratory tract specimens. A microorganism was isolated in blood cultures in 0.2% of cases (3/1157), two attributed to S. pneumoniae and one to S. aureus. The most prevalent bacteria isolated from respiratory tract specimens were H. influenzae (16.3%, 8/49), followed by S. pneumoniae (14.3%, 7/49), P. aeruginosa (14.3%, 7/49), S. aureus (14.3%, 7/49), and Escherichia coli (10.2%, 5/49). Five (11.4%, 5/49) mixed infections were detected (caused by Achromobacter xylosoxidans/Acinetobacter spp., S. pneumoniae/Enterobacter cloacae, E. coli/S. aureus, and Stenotrophomonas maltophilia/E. coli).
Main demographic information, patient characteristics, laboratory findings, treatments, and outcomes by respiratory groups are shown in Table 1. Patients with co-infection exhibited greater requirements for invasive mechanical ventilation (28.8% vs 19.0%, p = 0.035), prolonged hospital stays (14 vs 11 days, p = 0.003), elevated 30-day readmission rates (10.0% vs 4.2%, p = 0.025), and a higher prevalence of respiratory (23.8% vs 11.2%, p = 0.001) and urinary bacterial secondary infections (21.3% vs 10.6%, p = 0.004) (Supplementary table 1).
Predictors of respiratory bacterial co-infection
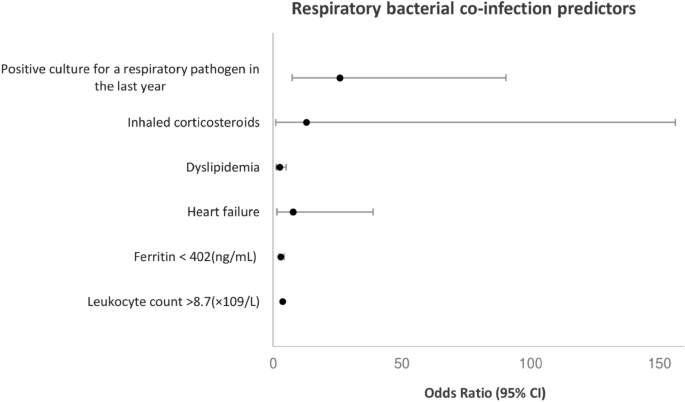
In the adjusted analyses (Fig. 1), respiratory bacterial co-infection was associated with having a positive culture for a respiratory pathogen in the last year (p < 0.001, OR = 25.89; 95% CI, 7.40–90.49), dyslipidaemia (p = 0.010, OR = 2.52; 95% CI, 1.25–5.08), heart failure (p = 0.015, OR = 7.68; 95% CI, 1.48–38.90), ferritin levels < 402 ng/mL (p = 0.011, OR = 2.28; 95% CI, 1.21–4.29), leukocyte count > 8.7 × 109/L (p = 0.004, OR = 2.40; 95% CI, 1.26–4.45), and patients with chronic obstructive pulmonary disease (COPD) treated with inhaled corticosteroids (ICS) (p = 0.044, OR = 12.94; 95% CI, 1.07–156.30).
Predictors of respiratory bacterial co-infection by multivariate logistic regression analyses. Culture for a respiratory pathogen in the last year (p < 0.001, OR = 25.89; 95% CI, 7.40–90.49), dyslipidemia (p = 0.010, OR = 2.52; 95% CI, 1.25–5.08), heart failure (p = 0.015, OR = 7.68; 95% CI, 1.48–38.90), ferritin levels < 402 ng/mL (p = 0.011, OR = 2.28; 95% CI, 1.21–4.29), leukocyte count > 8.7 × 109/L (p = 0.004, OR = 2.40; 95% CI, 1.26–4.45), and patients with chronic obstructive pulmonary disease (COPD) treated with inhaled corticosteroids (ICS) (p = 0.044, OR = 12.94; 95% CI, 1.07–156.30).
Empirical antimicrobial treatment at admission
Overall, 42.33% (n = 898/2121 patients with a positive SARS-CoV-2 NAAT) of eligible patients were treated with antibiotics. Antibiotic treatment rates were similar between patients with and without a bacterial co-infection (55.0% vs 56.5%, p = 0.788).
Ceftriaxone (81.1%, 729/898) and azithromycin (74.7%, 671/898) were the antibiotics most commonly used. The proportion of patients with antibiotics at admission decreased significantly across the different study periods (p < 0.001), as did the proportion of patients whose treatment was discontinued at 48 (p < 0.001) and 72 h (p < 0.001) post-admission (Supplementary table 2). In addition, the treatment duration, measured in DOT per 100 patient days, displayed an extended duration in the first period compared to the subsequent periods (Fig. 2).

Proportion of antibiotic use in hospitalised patients with COVID at admission (red), 48 (yellow) and 72 h (blue) post-admission, and Days of Therapy (DOT) per 100 patient days across different study periods (period 1: from March 2020 to May 2020; period 2: from June 2020 to December 9, 2020; period 3: from December 10 to March 15, 2021, and period 4: March 16, 2021, to May 1, 2021).
Impact of antibiotic treatment in patients admitted to ICUs
We observed no differences in mortality rates (55.0% vs 42.0%, p = 0.519), length of hospital stay (24 vs 22 days, p = 0.339), and 30-day readmission rates (3.0% vs 5.0%, p = 0.278) among ICU patients who had received antibiotics compared those who had not (Supplementary table 3). In addition, secondary infection rates (bacteremia: 21.8% vs 16.7%, p = 0.222; respiratory bacterial infection: 42.3% vs 44.7%, p = 0.648; urinary tract infection: 29.5% vs 34.0, p = 0.365), and percentages of multidrug-resistant microorganisms (ESBL: 9.5% vs 8.6%, p = 0.667; MRSA: 0.9% vs 2.6%, p = 0.192, XDR Pseudomonas aeruginosa: 2.3% vs 2.0 p = 0.813) did not differ between the two groups. Finally, the consumption of antibiotics, measured in DOT per 100 patient days, was higher in patients who had received antibiotics at admission (38.3 vs 26.6 days, p = 0.005) (Supplementary table 3).
Discussion
The current study provides a comprehensive characterization of respiratory bacterial co-infection, and the use and impact of antibiotics in clinical progression among hospitalised COVID-19 patients. Respiratory bacterial co-infection was documented in 6.9% of the patients, slightly higher than the percentage reported in a recent review, which described a co-infection rate of 4.4% among hospitalised patients2, 3, 7. This divergence could be attributed to the selection criteria of this study, in which patients were included if at least one urine sample for L. pneumophila and S. pneumoniae antigens was collected. In contrast, in other studies with lower co-infection rates, detection of S. pneumoniae antigen in urine was performed in fewer than 25% of patients3, 7. Conversely, a study conducted in Barcelona among hospitalized COVID-19 patients during the pandemic’s initial year reported a co-infection rate approaching 10%. The authors hypothesised that this elevated percentage could stem from diagnostic tests being requested solely in cases of clinical suspicion. Another explanation revolves around the cessation of lockdowns after the pandemic’s initial months, potentially facilitating increased community transmission of microorganisms like S. pneumoniae. Although the present study covers a similar period as the aforementioned study, most of the eligible patients in this study were from the pandemic’s outset (72.7% within the initial two study periods), which may not reflect the changes mentioned by Moreno-Garcia et al.8. This underscores the significance of tracking co-infection rates in forthcoming years.
While typically, community-acquired pneumonia (CAP) etiological agents, such as H. influenzae (16.3%, 8/49) and S. pneumoniae (14.3%, 7/49), accounted for many confirmed respiratory bacterial co-infections, healthcare-related microorganisms, such as P. aeruginosa (14.3%), S. aureus (14.3%) or E. coli (10.2%), were not negligible. The high percentage of patients with comorbidities (approximately 30% with COPD), prone to respiratory tract infections by these microorganisms, might account for their detection in this study.
In agreement with other studies3, 4, patients with bacterial co-infections displayed unfavourable outcomes. They required more invasive mechanical ventilation, experienced prolonged hospital stays, exhibited increased 30-day readmission rates, and were prone to secondary respiratory and urinary bacterial infections. Surprisingly, only 55% of patients with a respiratory bacterial co-infection received antibiotic treatment. Despite the limitations of this observational study, there is a clear need to improve the identification of patients with bacterial co-infection.
We were able to describe the predictors of respiratory bacterial co-infection, which is essential to design specific treatment protocols. To the best of our knowledge, this is the first study to establish links between having had a positive culture for a respiratory pathogen in the last year (OR = 25.89), dyslipidaemia (OR = 2.52), heart failure (OR = 7.68), and patients with COPD treated with ICS (OR = 12.94), with respiratory bacterial co-infection. In addition, Ferritin levels 8.7 × 109/L (OR = 2.4) were also associated with co-infection in adjusted analysis.
The presence of a positive culture for a respiratory pathogen in the preceding year exhibited a robust association with respiratory bacterial co-infection in this study. Previous works have also linked recent respiratory tract infections (caused by viruses and bacteria) to increased susceptibility for subsequent CAP9, 10.
Elevated leukocyte counts11, 12 and diminished ferritin levels8 have been previously associated with bacterial co-infection. Unlike other studies8, 13, we did not identify procalcitonin as an independent risk factor for bacterial co-infection. Although these biomarkers may not have sufficient sensitivity, specificity, or a positive predictive value by themselves, they can be combined with other clinical and radiological features to rule out bacterial co-infection with a high negative predictive value13, 14.
Dyslipidaemia has been linked to severe COVID-19 infections15 and higher rates of community-acquired sepsis16. Chronic inflammation and endothelial dysfunction15 may also facilitate bacterial co-infection in these patients. Moreover, as shown in the present study, chronic cardiovascular diseases, such as heart failure, have been described as a risk factor for CAP, and this association was to be expected17.
ICS are commonly employed to manage various chronic respiratory ailments like asthma or COPD, enhancing functionality, quality of life, and reducing exacerbation risks 18. However, ICS play a role in suppressing anti-bacterial host defences19 and increase the susceptibility to bacterial pneumonia20. This aspect could potentially favour the occurrence of bacterial co-infections among patients diagnosed with COVID-19.
As in other studies1, 7, the low percentage of patients with bacterial co-infection contrasts with the substantial number receiving antibiotics upon admission (42.33%). Nevertheless, the proportion of patients with antibiotics at admission decreased significantly in the different study periods, from 80.6% in the first study period (from March 2020 to May 2020) to 19.4% in the last study period (March 16, 2021, to May 1, 2021) (p < 0.001). This was also mirrored the patients whose treatment ceased at 48 and 72 h post-admission, probably due to the absence of data on bacterial co-infection, and the lack of local and international recommendations in the early stage of the pandemic. Furthermore, it was suggested that some antibiotics, such as azithromycin, could benefit in patients with COVID-19 due to their immunomodulatory properties, possibly driving over-treatment in these patients21.
Finally, we analysed the antibiotics use and their effects on a subset of ICU patients. We observed no differences in mortality, hospital stay duration, and 30-day readmission rates. Similarly, a study conducted in China found no association between early antibiotic use and mortality among non-severe COVID-19 patients admitted without bacterial infection. However, the patients treated with antibiotics were more at risk of progressing to severe illness and had protracted hospital stays. The authors suggested that antibiotic-induced dysbiosis might contribute to inflammation dysregulation in COVID-19 disease22.
Although we observed no differences in nosocomial infection rates and the prevalence of multidrug-resistant microorganisms (11.8% vs 14.0%, p = 0.616) between ICU patients who had received antibiotics at admission and those who had not, the DOT per 100 patient days was significantly greater in the first group (38.3 vs 26.6 days, p = 0.005). This could potentially wield significant consequences for the local ecology, potentially favouring the spread and outbreaks of high-risk clones23.
This study does exhibit some limitations. Despite encompassing an extensive study period (from March 2020 to May 2021), variations in bacterial co-infection rates and empirical antibiotic therapy may arise due to changes in clinical guidelines and the profile of patients admitted to the hospital, especially in terms of their severity. Secondly, the inclusion criteria involve patients who have been admitted for more than 48 h. This criteria excludes all patients who were discharged from the emergency department with antibiotic prescriptions, and it has the potential to favour the selection of patients with more severe clinical presentations. Thirdly, our definition of respiratory bacterial co-infection solely relies on retrospective positive cultures, rather than incorporating radiological or clinical features, potentially leading to both over and underdiagnosis. Nevertheless, the considerable number of patients in the study and the differences in septic and inflammatory parameters observed between the two groups strongly imply the presence of authentic bacterial co-infections. Fourthly, the previous use of antibiotics could have influenced the yield of bacterial cultures. However, we observed no significant differences between the co-infected (6.3%) and not co-infected groups (5.7%) in terms of the proportion of antibiotics administered prior to admission. Fifthly, we did not use non-culture tests, such as the nucleic acid amplification or serology assays, to detect respiratory pathogens. This could have potentially led to an underestimation of respiratory bacterial co-infection caused by pathogens such as Chlamydia pneumoniae and Mycoplasma pneumoniae24. However, a recent meta-analysis suggests that co-infections by these microorganisms were infrequent in COVID-19 patients1. Lastly, it’s important to note that this study was conducted at a single centre, and the incidence and epidemiology of respiratory bacterial co-infections may differ across geographic areas.
In conclusion, respiratory bacterial co-infection was low in a large cohort of hospitalised COVID-19 patients, although it could lead to worse outcomes in these patients. In addition, the percentage of patients with empirical antibiotic treatment remained high, even though it declined from the initial phase of the pandemic. Furthermore, empirical antibiotic treatment did not show exhibit benefits in patients admitted to ICUs. The present study allows identifying clinical and analytical predictors of respiratory bacterial co-infection within this population and could thereby help design targeted diagnostic and empirical antibiotic treatment protocols. A more comprehensive understanding of respiratory bacterial co-infection is essential to effectively identify high-risk patients and to implement suitable antibiotic stewardship measures.
Data availability
The study protocol, statistical analysis plan, and databases are available to anyone who requests them. These requests should be directed to Adrián Antuori ([email protected]).
References
-
Langford, B. J. et al. Bacterial co-infection and secondary infection in patients with COVID-19: A living rapid review and meta-analysis. Clin. Microbiol. Infect. 26(12), 1622–1629 (2020).
Google Scholar
-
Langford, B. J. et al. Predictors and microbiology of respiratory and bloodstream bacterial infection in patients with COVID-19: Living rapid review update and meta-regression. Clin. Microbiol. Infect. 28(6), 888–889 (2022).
Google Scholar
-
Garcia-Vidal, C. et al. Incidence of co-infections and superinfections in hospitalised patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 27(1), 83–88 (2021).
Google Scholar
-
Karaba, S. M. et al. Prevalence of co-infection at the time of hospital admission in COVID-19 Patients, a multicenter study. Open Forum Infect. Dis. 8(1), ofaa578 (2020).
Google Scholar
-
Pfister, R. et al. Procalcitonin for diagnosis of bacterial pneumonia in critically ill patients during 2009 H1N1 influenza pandemic: A prospective cohort study, systematic review and individual patient data meta-analysis. Crit. Care 18(2), R442014 (2014).
Google Scholar
-
Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18(3), 268–281 (2012).
Google Scholar
-
Karami, Z. et al. Few bacterial co-infections but frequent empiric antibiotic use in the early phase of hospitalised patients with COVID-19: Results from a multicentre retrospective cohort study in The Netherlands. Infect. Dis. (Auckl) 53(2), 102–110 (2021).
Google Scholar
-
Moreno-García, E. et al. Bacterial co-infection at hospital admission in patients with COVID-19: Bacterial co-infections in COVID-19. Int. J. Infect. Dis. 118, 197–202 (2022).
Google Scholar
-
Almirall, J. et al. Risk factors for community-acquired pneumonia in adults: A population- based case-control study. Eur. Respir. J. 13(2), 349–355 (1999).
Google Scholar
-
Hedlund, J. U. et al. Risk of pneumonia in patients previously treated in hospital for pneumonia. Lancet 340(8816), 396–397 (1992).
Google Scholar
-
Mason, C. Y. et al. Exclusion of bacterial co-infection in COVID-19 using baseline inflammatory markers and their response to antibiotics. J. Antimicrob. Chemother. 76(5), 1323–1331 (2021).
Google Scholar
-
Wang, L. et al. An observational cohort study of bacterial co-infection and implications for empirical antibiotic therapy in patients presenting with COVID-19 to hospitals in North West London. J. Antimicrob. Chemother. 76(3), 796–803 (2021).
Google Scholar
-
Carbonell, R., Moreno, G. M., Martín-Loeches, I., Bodí, M. & Rodríguez, A. The role of biomarkers in influenza and COVID-19 community-acquired pneumonia in adults. Antibiotics 12(1), 161 (2023).
Google Scholar
-
Vaughn, V. et al. Empiric antibacterial therapy and community-onset bacterial coinfection in patients hospitalised with coronavirus disease 2019 (COVID-19): A multi-hospital cohort study. Clin. Infect. Dis. 72(10), e533–e541 (2021).
Google Scholar
-
Hariyanto, T. I. & Kurniawan, A. Dyslipidemia is associated with severe coronavirus disease 2019 (COVID-19) infection. Diabetes Metab. Syndr. Clin. Res. Rev. 14(5), 1463–1465 (2020).
Google Scholar
-
Guirgis, F. W. et al. Cholesterol levels and long-term rates of community-acquired sepsis. Crit. Care 20(1), 408 (2016).
Google Scholar
-
Torres, A., Peetermans, W. E., Viegi, G. & Blasi, F. Risk factors for community-acquired pneumonia in adults in Europe: A literature review. Thorax 68(11), 1057–1065 (2013).
Google Scholar
-
Tashkin, D. P. & Strange, C. Inhaled corticosteroids for chronic obstructive pulmonary disease: What is their role in therapy?. Int. J. COPD 13, 2587–2601 (2018).
Google Scholar
-
Singanayagam, A. et al. Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci. Transl. Med. 11(507), eaav3879 (2019).
Google Scholar
-
Suissa, S., Patenaude, V., Lapi, F. & Ernst, P. Inhaled corticosteroids in COPD and the risk of serious pneumonia. Thorax 68(11), 1029–1036 (2013).
Google Scholar
-
Rosenberg, E. S. et al. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. JAMA 323(24), 2493–2502 (2020).
Google Scholar
-
Yin, X. et al. Evaluation of early antibiotic use in patients with non-severe COVID-19 without bacterial infection. Int. J. Antimicrob. Agents 59(1), 106462 (2022).
Google Scholar
-
O’Toole, R. F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 27(12), 1772–1776 (2021).
Google Scholar
-
Oliva, A. Co-infection of SARS-CoV-2 with Chlamydia or Mycoplasma pneumoniae: A case series and review of the literature. Infection 48(6), 871–877 (2020).
Google Scholar
Acknowledgements
We thank the COVID Unit of Laboratori Clínic Metropolitana Nord at Hospital Universitari Germans Trias i Pujol for their contribution in generating SARS-CoV-2 RT-PCR diagnostics data. This work was supported by the Ministry of Health of Government of Catalonia (Spain); AGAUR 2021 SGR 00931.
Author information
Authors and Affiliations
Contributions
Methodology: A.A., M.G.; Formal analysis and investigation: A.A.; Writing—original draft preparation: A.A., G.L.; Writing—review and editing: A.A., M.G., G.L., P.J.C.; Supervision: M.G., P.J.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Tables.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Antuori, A., Giménez, M., Linares, G. et al. Characterization of respiratory bacterial co-infection and assessment of empirical antibiotic treatment in patients with COVID-19 at hospital admission.
Sci Rep 13, 19302 (2023). https://doi.org/10.1038/s41598-023-46692-x
-
Received: 28 April 2023
-
Accepted: 03 November 2023
-
Published: 07 November 2023
-
DOI: https://doi.org/10.1038/s41598-023-46692-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.