Congenital disorders
Perspectives of current understanding and therapeutics of Diamond-Blackan anemia
Abstact
Diamond-Blackfan anemia (DBA) is a rare congenital bone marrow failure disorder characterized by erythroid hypoplasia. It primarily affects infants and is often caused by heterozygous allelic variations in ribosomal protein (RP) genes. Recent studies also indicated that non-RP genes like GATA1, TSR2, are associated with DBA. P53 activation, translational dysfunction, inflammation, imbalanced globin/heme synthesis, and autophagy dysregulation were shown to contribute to disrupted erythropoiesis and impaired red blood cell production. The main therapeutic option for DBA patients is corticosteroids. However, half of these patients become non-responsive to corticosteroid therapy over prolonged treatment and have to be given blood transfusions. Hematopoietic stem cell transplantation is currently the sole curative option, however, the treatment is limited by the availability of suitable donors and the potential for serious immunological complications. Recent advances in gene therapy using lentiviral vectors have shown promise in treating RPS19-deficient DBA by promoting normal hematopoiesis. With deepening insights into the molecular framework of DBA, emerging therapies like gene therapy hold promise for providing curative solutions and advancing comprehension of the underlying disease mechanisms.
Introduction
Diamond-Blackfan anemia (DBA) is a congenital bone marrow (BM) failure disorder with erythroid hypoplasia that presents early in infancy (5–7 cases per million live birth) [1]. The disease is also categorized as ribosomopathy [2, 3]. Around 75% of cases of DBA are related to a heterozygous allelic variation in ribosomal protein genes (RP) of either the small or large ribosomal subunit [4]. Until now, more than 20 RP genes have been identified. In addition, mutations in non-RP genes such as GATA1 and TSR2 were also identified as a cause of the DBA phenotype [4]. Hematopoietic stem cell transplantation is currently the sole curative option for the treatment of DBA [1]. This treatment is, however, limited by the availability of suitable donors and the potential for serious immunological complications. A recent study demonstrated that gene therapy using a clinically applicable lentiviral vector could rescue the impaired anemia in both mouse and human RPS19-deficient DBA models, with a low risk of mutagenesis and a highly polyclonal insertion site pattern, providing evidence for a potential curable treatment for patients with RPS19-deficiency [5]. In the present review, we discuss recent molecular and genetic understanding and new advancements in novel therapeutics for DBA.
History of the disease
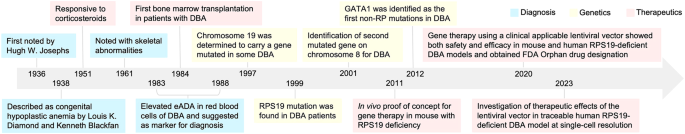
DBA was first reported by Hugh W. Josephs in 1936 [6], and more completely described by pediatricians Louis K. Diamond and Kenneth Blackfan who named the disorder as congenital hypoplastic anemia in 1938 [7] (Fig. 1). In 1951, corticosteroids were first reported to show therapeutic effects by Gasser [8], followed by a study of Diamond et al. indicating that a group of patients could respond to corticosteroid therapy [9]. In 1976, the first known bone marrow transplantation was performed on a 13-year-old boy with DBA who never responded to corticosteroid therapy and had received 238 transfusions, but iron chelation therapy showed no effects [10]. Initially, the treatment progressed well with erythroid precursors production was detected in the patient’s marrow for the first time in his life. However, the patient developed interstitial pneumonia and died 55 days after the transplant [10]. Elevated erythrocyte adenosine deaminase activity (eADA) in DBA patients was first reported and suggested as a marker for DBA by Diamond et al. in 1983 [11]. In 1997, a region on chromosome 19 was determined to carry a gene mutated in some DBA patients [12, 13]. Followed by this, mutations in the ribosomal protein S19 gene (RPS19) were found to be associated with disease in 42 of 172 DBA patients in 1999 [14]. Two years later, a second DBA gene was localized to a region of chromosome 8, and further genetic heterogeneity was inferred [15]. In 2012, the first non-RP gene, GATA1, was identified to have relationship with DBA, which broadened the understanding of molecular mechanism for DBA [16, 17]. The first in vivo prove-of-concept study by using gene therapy for the treatment of DBA was demonstrated in a mouse model with rps19 deficiency in 2011 [18]. Followed by gradual optimization of the therapeutic vector, a clinically applicable lentiviral vector where the RPS19 gene was driven by a cellular promoter, was shown to achieve both safety and efficacy in rescuing anemia and promote normal hematopoiesis in mouse and human RPS19-deficient models in 2021 [5]. Supported by this, the gene therapy strategy was approved for Orphan Drug Designation from FDA for further clinical trial investigation in patients with the RPS19 mutation. Our recent study further demonstrated the therapeutic effects of the vector in a traceable precise RPS19-deficient human DBA model at single-cell resolution [19, 20].
Timeline of understanding the history of DBA.
Clinical presentation
DBA is characterized by a paucity of erythroid progenitor and precursor cells in the bone marrow and red cell aplasia, and about half of the patients have physical malformations such as craniofacial defects, thumb deformities and short stature [4, 21]. Individuals with DBA also have a higher chance to develop cancer, including haematological malignancies (myelodysplastic syndrome, acute myeloid leukaemia) and solid tumors such as colon carcinoma and osteosarcomas [4, 21, 22].
Specifically, some DBA patients can also enter a state of remission [21, 23]. The DBA Registry defines “remission” as an adequate hemoglobin level without any treatment, lasting 6 months, independent of prior therapy [21]. The calculated likelihood of remission is 20% by age 25 years, with 72% experiencing a remission during the first decade of life [21]. Women also may relapse during pregnancy, with hormonal stress due to pregnancy appears to contribute to relapse [23, 24].
Diagnosis and genetic screening of DBA
A detailed discussion about diagnosis was well described by Jeffrey M. Lipton et al. [23]. Briefly, the classic laboratory presentations of DBA include severe anemia (macrocytic or normocytic) and reticulocytopenia present within the first year of life, further supported by absence or limited cytopenias of other lineages, and a visible paucity of erythroid precursor cells in the bone marrow [23]. However, not all the patients present with the classic clinical criteria, and cases diagnosed in adults were also described [25]. In addition, bone marrow aspiration is also used to distinguish from other hypogenerative anemia and bone marrow failure. Apart from these, the eADA activity is a useful diagnostic biomarker for diagnosis [11, 21], which is elevated in 80% to 85% DBA patients [4, 26, 27] and it usually remains elevated even in patients who are in remission or are hematologically stable with corticosteroids treatment [21]. Ulirsch et al. also observed a significant association where RPS19 and RPS24 individuals appear less likely to have elevated eADA in a cohort study [4].
Molecular analysis is also used to identify genetic lesions. Genetic screening starts with targeted Sanger sequencing of RPS19 (the most frequent genetic mutation) or directed next generation sequencing to analyse commonly mutated gene panels or all DBA related genes were applied according to the availability of the laboratory. Due to the limited incidence rate of the disease, DBA is not included in the universal prenatal screening for genetic disorders. However, when the DBA-causing pathogenic variant has been identified in an affected family member, it’s strongly advised to conduct prenatal testing for a pregnancy at increased risk and preimplantation genetic testing [21]. Details of DBA genetics will be discussed in the following paragraph.
Genetics of DBA
RP genes
Around 70–80% of the DBA cases were found to have putatively causal haploinsufficient variants in genes encoding proteins that comprise the large 60S (RPL) or small 40S (RPS) ribosomal subunit, suggesting that these mutations mainly reduce ribosome levels, leading to a selective reduction in the translation of key genes involved in erythroid lineage commitment during hematopoiesis [4, 22]. Up to now, mutations in 23 RP genes have been identified and are heterozygous, which inherited in an autosomal dominant pattern (Table 1). Homozygosity is largely suspected to be lethal, supported by the lethality of homozygous RP gene mutations in several animal models [28, 29]. Among these, RPS19, RPL5, RPS26, and RPL11 are the most frequently mutated RP genes [22]. A cohort study of 472 individuals with a clinical diagnosis of DBA showed that majority of the mutations are rare loss-of-function (LoF) alleles or missense, where 80% of mutations are a unique case [4]. Most putative causal mutations were typical LoF alleles or disrupted canonical mRNA splice sites, while the mutations predominately affect certain case of the extended consensus splice acceptor or donor site and a small number of rate mutations further from the exon-intron junction were also observed in the cohort [4]. Moreover, a mutation in the 3’UTR of RPS26 was also reported, which was predicted to completely disrupt the polyadenylation signal by changing the consensus motif AA(T/U)AAA to AAGAAA [4]. There are also 7 candidates RP genes were considered to have relationship with DBA, which are extremely intolerant to LoF mutation [4].
There is no strong relationship with any specific mutation gene for the specific syndrome. However, neutropenia is more frequently associated with RPL35a [30,31,32], cleft palate and abnormal thumbs with RPL5 and RPL11 [33]. Specifically, patients with RPL5 (83% on average) or RPL11 (73% on average) mutations had higher chance with one or more congenital malformations, compared with mutations in the RPS19 gene (34% on average) [4, 34]. Patients with RPS24 (36%) and RPL11 (29%) have higher chance to develop remission, compared with RPS19 (8%) and RPL5 (5%) mutations [4]. There is no significant difference in the treatment requirements for transfusion or corticosteroid dependence among mutations in the RP genes according to current experience [4].
Non-RP genes
In 2012, GATA1 was identified as the first non-RP mutation in DBA using whole exome analysis [17]. GATA1 is a hematopoietic master transcription factor that is both necessary for proper erythropoiesis and sufficient to reprogram alternative hematopoietic lineages to an erythroid fate [35]. The mutations were found at a splice donor site of the GATA1 gene, and this leading to the impaired production of the full-length form of the protein, which required for normal erythropoiesis in humans [17, 36]. In addition, 2 RP chaperones, TSR2 [37] and HEATR3 [38], have also been identified in DBA patients. The ribosomal assembly factor TSR2, which is an RPS26 chaperone (X-chromosomal gene encoding a direct binding partner of RPS26), has a critical role in ensuring adequate ribosome levels in hematopoietic progenitors [39]. Several individuals present with biallelic variants in HEATR3 were shown to have association with DBA [38]. The HEATR3 variants destabilize the protein, resulting in a reduction of nuclear uL18 (RPL5) and impaired ribosome biogenesis independent of p53 in CD34+ cells [38]. In particular, individuals with HEATR3 variants exhibit more severe phenotype with bone marrow failure, short stature, facial and acromelic dysmorohic feature, and intellectual disability [38]. Specifically, GATA1-related and TSR2-related DBA are inherited in an X-linked manner, and HEATR3 is inherited in a recessive manner [17, 38].
Moreover, EPO [40] and CECR1 [41] were shown to be the DBA-associated genes. A homozygous recessive mutation in EPO (R150Q) was reported in an individual, and the mutation shows a mild reduction in affinity for its receptor but also altered binding kinetics, leading to less effective at stimulating erythroid cell proliferation and differentiation [40]. The cohort study identified recessive CECR1 mutations in several individuals [41]. Each of the individuals presented with severe normocytic or microcytic anemia and bone marrow erythroid hypoplasia in infancy without any additional physical abnormalities. However, no abnormal rRNA maturation (typical in RP gene DBA) was observed in whole blood from 2 unrelated CECR1 individuals. And these individuals were not observed to have elevated eADA [4]. Because of this, mutations in CECR1 was regarded as DBA-like diseases, but screening for CECR1 is highly recommended when individuals present with DBA [22].
Molecular mechanisms of DBA
The pathophysiology of DBA has not been fully understood. Since many mutations are RP genes, the mainly unsolved question is how the mutation in an RP gene leading to an aberrant ribosome assembly and impaired ribosomal biogenesis leads to the impaired erythroid defect [42]. Translation regulation, p53 stabilization and cell cycle arrest, unbalanced globin/heme synthesis and autophagy were demonstrated to have relationship with DBA. Emerging evidence also indicated that inflammatory mechanisms may play a role in DBA (Fig. 2).

A p53 activation and cell cycle arrest leading to ribosomal stress. B Translational dysfunction caused by GATA1 and RP mutations. C Abnormal inflammatory signaling pathways due to RP mutations. D Unbalanced globin/heme synthesis caused by RP mutations.
p53 activation and cell cycle arrest
Ribosomal stress was known to inhibit p53 ubiquitination and induce p53 transactivation, which leads to p53-dependent cell cycle arrest and apoptosis [43, 44]. Many RPs involved in the regulation of p53 via interaction with its transcriptional target, MDM2, where RPs inhibit MDM2-medicated p53 proteasomal degradation [45]. Several RP-mutations in DBA have been observed with activations of p53 and target genes (especially RPL5 and RPL11) in both animal models and patient samples [46,47,48,49,50,51,52]. By analysing the differentiation trajectories from megakaryocytic-erythroid progenitors (MEPs) to red blood cells and platelets, Lu et al. demonstrated that knockdown of p53 leads to the reduction of MEPs and increase of erythroid progenitors [53, 54]. They also demonstrated that high cell cycle speed was required during MEPs fate decision, and erythroid progenitors have significantly more proliferation than megakaryocyte-committed progenitors by scRNA-seq analysis [54]. In addition, individuals with gain-of-function mutations in exon 10 of p53 gene were reported to have DBA-like syndromes between DBA and dyskeratosis congenita [55]. GATA1 was also demonstrated to have impact on p53 inhibition [56]. All these findings indicated the essential role of RP-mutations in the induction of p53 activation in the pathophysiology of DBA.
Translational dysfunction
Several studies of RP mutations have indicated at least modest reductions in overall protein synthesis [57]. It suggested that one main possibility is impaired translation of global or specific mRNAs in certain tissue leads to the specific ribosomopathy phenotype [36, 42, 58, 59]. The reduced RP expression was also known to lead to aberrant ribosome assembly and reduced ribosome levels. In most cases, the global protein synthesis is modestly reduced [42]. GATA1 is the master hematopoietic transcription factor of megakaryopoiesis and erythropoiesis [60]. Mutation in the splice donor site of GATA1 reduces the levels of full-length GATA1 protein and can cause DBA in certain individuals [17, 36]. In addition, in patients with RP-mutation DBA, GATA1 mRNA is poorly translated as the result of a highly structured 5’ untranslated region (5’UTR), and target genes of GATA1 also showed globally and specifically reduction, which indicated the activity reduction of GATA1 [36, 61]. It is still unknown how it impacts the reduction of GATA1 mRNA translation, one possibility maybe the requirement of higher threshold for initiation of translation of GATA1 mRNA compared to other genes [36, 62, 63].
Increased Inflammatory signaling pathway
Inflammatory signals are known to play a role for erythropoiesis. Overproduction of proinflammatory cytokines were shown to inhibit steady-state bone marrow erythropoiesis [64,65,66,67]. In contrast, inflammatory signals were demonstrated to induce stress erythropoiesis to maintain erythroid hemeostasis [68, 69]. Recent studies indicated inflammatory signatures would make impact on DBA, which may lead to the stress erythropoiesis. Elevated IFN-γ and TNF-α can be detected in DBA bone marrow plasma, and inflammatory signature was shown in erythroblasts and RBCs from DBA patients [70, 71]. By performing single cell RNA-seq (scRNA-seq) analysis using patient bone marrow HSPCs, increased IFN- α, IFN- γ, and TNF-α inflammatory pathways were identified in both RPS-DBA and RPL-DBA, with more obvious changes in RPS-DBA than RPL-DBA [70]. A previous reported zebrafish RPL11 morpholinos also indicated increased inflammation [72]. Moreover, in patients responding to glucocorticoids treatment, increased type 1 interferon pathway was found to inhibit cell cycle progression by scRNA-seq analysis [73]. Interestingly, a low dose of interferon alpha treatment could promote RBC production in cells isolated from DBA [73]. Our recent study also identified enrichment of TNFα/NF-κB in gene edited human RPS19-deficient CD34+ cells by scRNA-seq analysis, while this was not observed in RPS19-deficient CD34+ cells treated with clinical applicable lentiviral vector [19]. The inflammatory signature also provides possible mechanism on how glucocorticoids exert their therapeutics in DBA [74, 75]. Taken together, both cell intrinsic and extrinsic defects may trigger inflammatory responses. Future studies about how inflammatory pathways contribute to the disease are worth to be explored.
Unbalanced globin/heme synthesis
The imbalance in excess free heme, which leads to production of reactive oxygen species were shown toxic to cells and leads to cell death and apoptosis [76, 77]. Similar to this, imbalanced globin and heme synthesis in primary DBA cells have been reported to lead to accumulation of free heme and heme toxicity in early erythroid precursors, which perturbs erythroid differentiation [78, 79]. In addition, mice with knockout of heme exporter, feline leukemia virus subgroup C receptor (FLVCR1), display impairment of erythropoiesis and congenital abnormalities as observed in DBA patients [80]. In some DBA patients negative for RPS19 gene mutations, alternatively spliced isoforms of FLVCR1 were also identified in immature bone marrow erythroid cells [81]. A recent study also found elevated FLVCR1 expression in patients with RPL11 and RPL5 mutations, and decreased GATA1 was also observed meanwhile [78]. Since α and β globins are transcriptionally regulated by GATA1, the reduction of GATA1 was hypothesized leading to the imbalance heme/globin [82]. As HSP70 is subjected to proteasomal degradation leading to decreased levels of GATA1 in erythroid cells with RPL5 and RPL11 mutations [83], the author further demonstrated that overexpression of HSP70 could protect GATA1 and restore heme/globin balance. These findings imply the role of FLVCR1 in the regulation of human erythroid cells through control of the heme content, which induce apoptosis on erythroid cells.
Autophagy
Autophagy is an important catabolic process that delivers cytoplasmic material to the lysosome for degradation. It promotes cell survival by elimination of damaged organelles and proteins aggregates, as well as by facilitating bioenergetic homeostasis [84]. A small molecular act through autophagy factor ATG5 was identified to promote erythropoiesis and up-regulate expression of globin genes in induced pluripotent stem cells isolated from DBA patients and in vivo [85]. How the autophagy was regulated in DBA is not well understood. Autophagy was shown to affect erythropoiesis through degradation of the iron storage protein ferritin [86, 87]. And Atg5-deficient zebrafish are anemic, indicating that ATG5 plays a role in erythroid development. More detailed studies are necessary to demonstrate the mechanism of autophagy on DBA.
Treatments
Glucocorticoids, management and side effects
Glucocorticoids (GC) are the only widely used class of drugs in DBA since their first report in 1950s [8], about 80% patients respond to the therapeutic at the beginning, while half of these patients eventually discontinue GC treatment due to loss of response or severe side effects, such as growth retardation, pregnancy, etc [1, 21]. Until now, DBA is the only human disease in which steroids are administrated for years. Treatment with GC is not recommended in patients less than 1 year old due to growth inhibition [1, 88, 89]. The adequate response is defined as a hemoglobin level >90 g/l in combination with transfusion independency [1]. In general, treatment with GC is started with an initial dose of 2 mg/kg/day prednisone for a maximal period of 4 weeks [1]. In case of a response, slow tapering (below 1 mg/kg/day) is indicated to the lowest effective doses after initial 4 weeks [1, 21]. In most guidelines, 0.3–0.5 mg/kg/day of prednisone is regarded as the highest acceptable level to avoid long term toxicities [1, 22]. It is also recommended to take vitamin D supplementation to all DBA patients and perform periodic bone density measurements [1]. About 40% of case subjects remain dependent upon corticosteroids which increase the risk of heart disease, osteoporosis, and severe infections [1, 23]. For patients who make no or limited response on reticulocytes and hemoglobin levels, blood transfusion or hematopoietic stem cell transplantation are considered.
The mechanism of how GC works still not well understood and under investigation. A detailed review about the relationships of GCs on DBA through interacting with GATA1, p53, c-myc, mTOR and autophagy were well described by Zuzana Macečková et al. [90]. In untreated DBA patients, Wang et al. recently showed that erythroid progenitors entered S-phase of the cell cycle under considerable stress, leading to replication stress and activation of p53 signaling [73]. However, in GC-responsive patients, cell cycle progression was inhibited by activation of the type 1 interferon pathway compared with GC-non-responsive patients [73]. Moreover, Iskander et al. also showed that the stress erythropoiesis in RPL-DBA exhibited disordered differentiation by an altered glucocorticoid molecular signature, including reduced ZFP36L2 expression, leading to milder anemia and improved corticosteroid response compared with RPS-DBA [70]. In addition to this, Ryan et al. also demonstrated that dexamethasone treatment of peripheral blood progenitors can result in the expansion of a newly defined immature colony-forming unit (CD34+CD36+CD71hiCD105med) by activation of p57Kip2, which is a Cip/Kip cyclin-dependent kinase inhibitor. Notably, steroid resistance was shown to be associated with dysregulated p57Kip2 expression. In particular, this only happened in peripheral blood, not cord blood [91]. Taken together, a complex mechanism involving translation, proliferation and differentiation may all together contribute to the GC response.
Transfusion and management of iron overload
For patients who do not response to corticosteroid treatment have to be given blood transfusions [1, 21, 23]. Basically, patients require 10–15 ml/kg per RBC transfusion every 3–5 weeks to maintain hemoglobin levels above 80 g/l [1, 21, 23]. For infants and young children, higher levels of hemoglobin (>90 g/l) are required to maintain adequate growth and development [1]. However, the toxicity associated with iron overload, concomitant with chronic transfusion regimens, is a limiting factor for lifelong transfusion [1, 21]. The transfusion-associated iron overload is a leading cause of mortality in DBA patients in addition to HSCT-related mortality [1, 92]. Because of this, the effective and intense chelation therapy is necessary for DBA patients. Currently, the best and most feasible way to analyse iron overload is to perform magnetic resonance imaging (MRI)-based measurements of hepatic, cardiac and pancreatic iron burden [1]. If MRI is not available or applicable, serum ferritin levels of ≥1000 ug/l and/or transferrin saturation levels ≥75% are considered as a starting point for chelation therapy [1]. It is recommended to measure liver iron content every 12–18 months on chronic RBC transfusion treatment or screen for iron overload and start chelation therapy after 10–20 RBC transfusion (of 10–15 ml/kg), or when the MRI-measured liver iron concentration reaches ≥6–7 mg/g [1, 23]. Chelators such as deferoxamine or combination with deferasirox are used to achieve ferritin levels less than 500 ug/L and normal liver iron status by magnetic resonance imaging [22, 93, 94]. Specifically, deferoxamine is more preferred to use for infants as with supporting data [22].
Hematopoietic stem cell transplantation, managements and side effects
Currently, hematopoietic stem cell transplantation (HSCT) is the only curative treatment for DBA [1, 21, 22]. Standard indications for HSCT include resistance to GC treatment, chronic transfusion dependency and unacceptable GC toxicity [1, 95]. Recent studies suggest that HSCT should be recommended for transfusion-dependent children aged less than 10 years who make no response to GC or require high doses if a human leucocyte antigen (HLA) matched donor is available [1, 96,97,98,99]. The HLA-matched family donors are preferred donor type, and genetic screening of the affected gene for DBA to avoid an asymptomatic DBA carrier donor is necessary in cases with a known underlying genetic lesion. For patients with no mutation could be identified, it’s essential to assess potential related donors through erythrocyte adenosine deaminase analysis and a bone marrow test to exclude a silent carrier [98]. However, if this is not available, a 10/10 allele-matched unrelated donor is the best alternative [1]. For stem cell source, stem cells from bone marrow are more preferred than from peripheral blood due to lower risk of chronic graft versus host disease (GVHD) [96]. Umbilical cord blood derived stem cells from a sibling donor can also be considered if available, while transplantation with unrelated donors showed higher graft failure and transplant-related mortality rates [96]. For conditioning regimens, total body irradiation should be avoided in infants and not recommended for other DBA patients as it increases the risk of secondary malignancies as they already have higher risk for cancer predisposition. Myeloablative conditioning with busulfan, and more recently treosulfan, has been recommended as a means of favouring engraftment and reducing graft failure [96]. In recent years, low dose conditioning was suggested and has been demonstrated with efficacy in clinical trial [100, 101]. Our group also demonstrated the full correction of the hematopoietic phenotype in DBA mice given sublethal doses of irradiation, as well as in animals completely devoid of any proceeding irradiation [102]. In addition to this, antibody approach also showed promising effects with less toxic effects compared with conditioning [103, 104]. Donor rejection and GVHD also need to be considered when perform HSCT [1, 96]. Sufficient immunoablation (eg. Fludarabine) and serotherapy showed effects in reducing the risk of graft rejection and GVHD, especially for patients receiving unrelated donor [96, 98]. In addition, infertility is also a major concern after transplantation. Counselling about fertility preservation before transplantation and post-transplant follow-up are recommended [1, 96, 105].
Gene therapy, safety management and future perspectives
Gene therapy using genetically engineered human hematopoietic stem and progenitor cells (HSPCs) is a potential therapeutic strategy for genetic blood disorders [106, 107] (Fig. 3). The use of self-inactivating lentiviral vectors for ex vivo gene correction of HSPCs has been successfully applied to treat primary immunodeficiencies [108, 109], haemoglobinopathies [110, 111] and metabolic disorders [112, 113] with superior engraftment and safer profile in patients [106, 107]. We recently demonstrated gene therapy using a clinically applicable lentiviral vector driven by a cellular promoter, EFS, could promote red blood cell production and normal hematopoiesis in a mouse DBA model with RPS19 deficiency and human RPS19-deficient CD34+ HSPCs, with a low risk of mutagenesis and a highly polyclonal insertion site pattern [114]. Followed by this, similar strategies also showed rescue effects by other groups [115]. In addition, using lentiviral vector to express GATA1 so that to promote red blood production is also being investigated, which also provides advantages for targeting most DBA mutations instead of a specific mutation [116]. However, the control of GATA1 expression level is worth to be carefully investigated considering its regulation function as a transcription factor.

Summary of therapeutic alternatives for DBA.
The successful development of gene therapy for RPS19-deficient DBA opens the possibilities for other mutations of DBA, such as RPL5 and RPL11. In addition, gene therapy using CRISPR-Cas9 genome editing tools also showed therapeutic effects for genetic blood disorders such as sickle cell disease and beta-thalassemia [117,118,119]. The CRISPR-Cas9 derived editing tools such as high-fidelity (HiFi) Cas9, base editors and prime editors could improve editing efficiency with reduced off-targets or without double-strand DNA cleavage [119, 120]. These genome editing tools provide possibilities to directly edit mutated genes by using base editors [117], or edit erythroid-specific enhancer region of BCL11A with CRISPR-Cas9 [118], or deliver a full-length therapeutic gene site specifically using HiFi Cas9 and AAV via homologous recombination in HSPCs for hematologic disorders [119]. The above strategies can also be considered to develop gene therapy for DBA in the future. However, since p53 activation was observed in patients with DBA, genotoxic risks are warranted to be considered when using gene editing [121, 122].
In addition to the ex vivo HSPC genetic manipulation, recent studies also demonstrated the possibility by using in vivo priming editing for the treatment of genetic blood disorder such as sickle cell disease in a mouse model [123], which provides significant advantages compared to the ex vivo gene therapy considering the needs of transplantation, ex vivo HSCs collection and myeloablative conditioning. This also opens the possibility for the development of in vivo gene therapy strategy for DBA. However, specific targeting to desired cell type such as HSPCs is essential to avoid off-targeting when performing in vivo delivery, which is also under investigation by using different delivery strategies such as viral vectors, lipid nanoparticles and virus-like particles [124].
Conclusion
Followed by the first clinical report of DBA in the 1930s, a better understanding of the diagnosis, genetics, molecular mechanisms and novel therapeutics of DBA has been made through working together by patients, families, clinicians and researchers. With the recent advancement of next generation sequencing, more RP and non-RP genes were found to have relationship with DBA, which helps with clinical diagnosis and provides new clues to discover molecular mechanisms. The successful development of the mouse and human DBA models also provide support for the investigation of mechanisms and novel therapeutics. In terms of therapeutics, autologous gene corrected HSPCs using clinically applicable lentiviral vector in animal models showed curative treatment potential with both safety and efficacy, which also avoids challenges such as GVHD and donor limitation compared to HSCT. The rapid evolution of genome-editing and delivery technologies also provides opportunities to precisely correct mutations in DBA in the future. However, the molecular mechanism of DBA is still not fully understood, and novel therapeutics such as gene therapy should also be developed for other mutations of DBA. Future attempts in the investigation of these aspects will bring better understanding and more therapeutic alternatives for DBA.
References
-
Bartels M, Bierings M. How I manage children with Diamond-Blackfan anaemia. Br J Haematol. 2019;184:123–33.
Google Scholar
-
Kang J, Brajanovski N, Chan KT, Xuan J, Pearson RB, Sanij E. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct Target Ther. 2021;6:323.
Google Scholar
-
Dianzani I, Loreni F. Diamond-Blackfan anemia: a ribosomal puzzle. Haematologica. 2008 ;93:1601–4.
Google Scholar
-
Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS, et al. The genetic landscape of Diamond-Blackfan anemia. Am J Hum Genet. 2018;103:930–47.
Google Scholar
-
Liu Y, Dahl M, Debnath S, Rothe M, Smith EM, Grahn THM, et al. Successful gene therapy of Diamond-Blackfan anemia in a mouse model and human CD34(+) cord blood hematopoietic stem cells using a clinically applicable lentiviral vector. Haematologica. 2022;107:446–56.
Google Scholar
-
Josephs HW. Anaemia of infancy and early childhood. Medicine. 1936;15:307–451.
-
Louis K, Diamond KB. Hypoplastic anemia. Am J Dis Child. 1938;56:464–7.
-
Gasser C. [Aplastic anemia (chronic erythroblastophthisis) and cortisone]. Schweiz Med Wochenschr. 1951;81:1241–2.
Google Scholar
-
Allen DM, Diamond LK. Congenital (erythroid) hypoplastic anemia: cortisone treated. Am J Dis Child. 1961;102:416–23.
Google Scholar
-
August CS, King E, Githens JH, McIntosh K, Humbert JR, Greensheer A, et al. Establishment of erythropoiesis following bone marrow transplantation in a patient with congenital hypoplastic anemia (Diamond-Blackfan syndrome). Blood. 1976;48:491–8.
Google Scholar
-
Glader BE, Backer K, Diamond LK. Elevated erythrocyte adenosine deaminase activity in congenital hypoplastic anemia. N Engl J Med. 1983;309:1486–90.
Google Scholar
-
Gustavsson P, Willing TN, van Haeringen A, Tchernia G, Dianzani I, Donner M, et al. Diamond-Blackfan anaemia: genetic homogeneity for a gene on chromosome 19q13 restricted to 1.8 Mb. Nat Genet. 1997;16:368–71.
Google Scholar
-
Gustavsson P, Skeppner G, Johansson B, Berg T, Gordon L, Kreuger A, et al. Diamond-Blackfan anaemia in a girl with a de novo balanced reciprocal X;19 translocation. J Med Genet. 1997;34:779–82.
Google Scholar
-
Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–75.
Google Scholar
-
Gazda H, Lipton JM, Willig TN, Ball S, Niemeyer CM, Tchernia G, et al. Evidence for linkage of familial Diamond-Blackfan anemia to chromosome 8p23.3-p22 and for non-19q non-8p disease. Blood. 2001;97:2145–50.
Google Scholar
-
Klar J, Khalfallah A, Arzoo PS, Gazda HT, Dahl N. Recurrent GATA1 mutations in Diamond-Blackfan anaemia. Br J Haematol. 2014;166:949–51.
Google Scholar
-
Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122:2439–43.
Google Scholar
-
Jaako P, Flygare J, Olsson K, Quere R, Ehinger M, Henson A, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118:6087–96.
Google Scholar
-
Liu Y, Schmiderer L, Hjort M, Lang S, Bremborg T, Rydstrom A, et al. Engineered human Diamond-Blackfan anemia disease model confirms therapeutic effects of clinically applicable lentiviral vector at single-cell resolution. Haematologica. 2023;108:3095–109.
-
Voit RA, Corey SJ. Gene therapy for congenital marrow failure syndromes – no longer grasping at straws? Haematologica. 2023;108:2880–2882.
-
Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116:3715–23.
Google Scholar
-
Da Costa L, Leblanc T, Mohandas N. Diamond-Blackfan anemia. Blood. 2020;136:1262–73.
Google Scholar
-
Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, Ramenghi U, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142:859–76.
Google Scholar
-
Faivre L, Meerpohl J, Da Costa L, Marie I, Nouvel C, Gnekow A, et al. High-risk pregnancies in Diamond-Blackfan anemia: a survey of 64 pregnancies from the French and German registries. Haematologica. 2006;91:530–3.
Google Scholar
-
Flores Ballester E, Gil-Fernandez JJ, Vazquez Blanco M, Mesa JM, de Dios Garcia J, Tamayo AT, et al. Adult-onset Diamond-Blackfan anemia with a novel mutation in the exon 5 of RPL11: too late and too rare. Clin Case Rep. 2015;3:392–5.
Google Scholar
-
Fargo JH, Kratz CP, Giri N, Savage SA, Wong C, Backer K, et al. Erythrocyte adenosine deaminase: diagnostic value for Diamond-Blackfan anaemia. Br J Haematol. 2013;160:547–54.
Google Scholar
-
Glader BE, Backer K. Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases. Br J Haematol. 1988;68:165–8.
Google Scholar
-
Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leveen P, et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24:4032–7.
Google Scholar
-
Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139.
Google Scholar
-
Gianferante MD, Wlodarski MW, Atsidaftos E, Da Costa L, Delaporta P, Farrar JE, et al. Genotype-phenotype association and variant characterization in Diamond-Blackfan anemia caused by pathogenic variants in RPL35A. Haematologica. 2021;106:1303–10.
Google Scholar
-
Noel CB. Diamond-Blackfan anemia RPL35A: a case report. J Med Case Rep. 2019;13:185.
Google Scholar
-
Tamefusa K, Muraoka M, Washio K, Wakamatsu M, Shimada A. Late-onset familial Diamond-Blackfan anemia with neutropenia caused by RPL35A variant. Pediatr Int. 2022;64:e15275.
Google Scholar
-
Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–80.
Google Scholar
-
Quarello P, Garelli E, Carando A, Cillario R, Brusco A, Giorgio E, et al. A 20-year long term experience of the Italian Diamond-Blackfan Anaemia Registry: RPS and RPL genes, different faces of the same disease? Br J Haematol. 2020;190:93–104.
Google Scholar
-
Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25:1215–27.
Google Scholar
-
Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20:748–53.
Google Scholar
-
Gripp KW, Curry C, Olney AH, Sandoval C, Fisher J, Chong JX, et al. Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A. 2014;164A:2240–9.
Google Scholar
-
O’Donohue MF, Da Costa L, Lezzerini M, Unal S, Joret C, Bartels M, et al. HEATR3 variants impair nuclear import of uL18 (RPL5) and drive Diamond-Blackfan anemia. Blood. 2022;139:3111–26.
Google Scholar
-
Yang YM, Karbstein K. The chaperone Tsr2 regulates Rps26 release and reincorporation from mature ribosomes to enable a reversible, ribosome-mediated response to stress. Sci Adv. 2022;8:eabl4386.
Google Scholar
-
Kim AR, Ulirsch JC, Wilmes S, Unal E, Moraga I, Karakukcu M, et al. Functional selectivity in cytokine signaling revealed through a pathogenic EPO mutation. Cell. 2017;168:1053–64.e1015.
Google Scholar
-
Szvetnik EA, Klemann C, Hainmann I, O’ Donohue M-F, Farkas T, Niewisch M, et al. Diamond-Blackfan anemia phenotype caused by deficiency of adenosine deaminase 2. Blood. 2017;130:874.
-
Mills EW, Green R. Ribosomopathies: there’s strength in numbers. Science. 2017;358:eaan2755.
-
Liu Y, Deisenroth C, Zhang Y. RP-MDM2-p53 pathway: linking ribosomal biogenesis and tumor surveillance. Trends Cancer. 2016;2:191–204.
Google Scholar
-
Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199–210.
Google Scholar
-
Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–77.
Google Scholar
-
Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–37.
Google Scholar
-
Moniz H, Gastou M, Leblanc T, Hurtaud C, Cretien A, Lecluse Y, et al. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3:e356.
Google Scholar
-
Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One. 2009;4:e4152.
Google Scholar
-
Torihara H, Uechi T, Chakraborty A, Shinya M, Sakai N, Kenmochi N. Erythropoiesis failure due to RPS19 deficiency is independent of an activated Tp53 response in a zebrafish model of Diamond-Blackfan anaemia. Br J Haematol. 2011;152:648–54.
Google Scholar
-
Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010;116:2826–35.
Google Scholar
-
McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–70.
Google Scholar
-
Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–76.
Google Scholar
-
Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–7.
Google Scholar
-
Lu YC, Sanada C, Xavier-Ferrucio J, Wang L, Zhang PX, Grimes HL, et al. The molecular signature of megakaryocyte-erythroid progenitors reveals a role for the cell cycle in fate specification. Cell Rep. 2018;25:3229.
Google Scholar
-
Fedorova D, Ovsyannikova G, Kurnikova M, Pavlova A, Konyukhova T, Pshonkin A, et al. De novo TP53 germline activating mutations in two patients with the phenotype mimicking Diamond-Blackfan anemia. Pediatr Blood Cancer. 2022;69:e29558.
Google Scholar
-
Trainor CD, Mas C, Archambault P, Di Lello P, Omichinski JG. GATA-1 associates with and inhibits p53. Blood. 2009;114:165–73.
Google Scholar
-
McGowan KA, Mason PJ. Animal models of Diamond Blackfan anemia. Semin Hematol. 2011;48:106–16.
Google Scholar
-
Horos R, Ijspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, Taskesen E, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–72.
Google Scholar
-
Gilles L, Arslan AD, Marinaccio C, Wen QJ, Arya P, McNulty M, et al. Downregulation of GATA1 drives impaired hematopoiesis in primary myelofibrosis. J Clin Invest. 2017;127:1316–20.
Google Scholar
-
Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–44.
Google Scholar
-
Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018;173:90–103.e119.
Google Scholar
-
Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–83.
Google Scholar
-
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42.
Google Scholar
-
Libregts SF, Gutierrez L, de Bruin AM, Wensveen FM, Papadopoulos P, van Ijcken W, et al. Chronic IFN-gamma production in mice induces anemia by reducing erythrocyte life span and inhibiting erythropoiesis through an IRF-1/PU.1 axis. Blood. 2011;118:2578–88.
Google Scholar
-
Rusten LS, Jacobsen SE. Tumor necrosis factor (TNF)-alpha directly inhibits human erythropoiesis in vitro: role of p55 and p75 TNF receptors. Blood. 1995;85:989–96.
Google Scholar
-
Xiao W, Koizumi K, Nishio M, Endo T, Osawa M, Fujimoto K, et al. Tumor necrosis factor-alpha inhibits generation of glycophorin A+ cells by CD34+ cells. Exp Hematol. 2002;30:1238–47.
Google Scholar
-
Zamai L, Secchiero P, Pierpaoli S, Bassini A, Papa S, Alnemri ES, et al. TNF-related apoptosis-inducing ligand (TRAIL) as a negative regulator of normal human erythropoiesis. Blood. 2000;95:3716–24.
Google Scholar
-
Bennett LF, Liao C, Quickel MD, Yeoh BS, Vijay-Kumar M, Hankey-Giblin P, et al. Inflammation induces stress erythropoiesis through heme-dependent activation of SPI-C. Sci Signal. 2019;12:eaap7336.
-
Paulson RF, Hariharan S, Little JA. Stress erythropoiesis: definitions and models for its study. Exp Hematol. 2020;89:43–54.e42.
Google Scholar
-
Iskander D, Wang G, Heuston EF, Christodoulidou C, Psaila B, Ponnusamy K, et al. Single-cell profiling of human bone marrow progenitors reveals mechanisms of failing erythropoiesis in Diamond-Blackfan anemia. Sci Transl Med. 2021;13:eabf0113.
Google Scholar
-
Kapralova K, Jahoda O, Koralkova P, Gursky J, Lanikova L, Pospisilova D, et al. Oxidative DNA damage, inflammatory signature, and altered erythrocytes properties in Diamond-Blackfan anemia. Int J Mol Sci. 2020;21:9652.
-
Danilova N, Wilkes M, Bibikova E, Youn MY, Sakamoto KM, Lin S. Innate immune system activation in zebrafish and cellular models of Diamond Blackfan Anemia. Sci Rep. 2018;8:5165.
Google Scholar
-
Wang B, Wang C, Wan Y, Gao J, Ma Y, Zhang Y, et al. Decoding the pathogenesis of Diamond-Blackfan anemia using single-cell RNA-seq. Cell Discov. 2022;8:41.
Google Scholar
-
Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol Cell Endocrinol. 2011;335:2–13.
Google Scholar
-
Cain DW, Cidlowski JA. Immune regulation by glucocorticoids. Nat Rev Immunol. 2017;17:233–47.
Google Scholar
-
Kumar S, Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol Lett. 2005;157:175–88.
Google Scholar
-
Gbotosho OT, Kapetanaki MG, Kato GJ. The worst things in life are free: the role of free heme in sickle cell disease. Front Immunol. 2020;11:561917.
Google Scholar
-
Rio S, Gastou M, Karboul N, Derman R, Suriyun T, Manceau H, et al. Regulation of globin-heme balance in Diamond-Blackfan anemia by HSP70/GATA1. Blood. 2019;133:1358–70.
Google Scholar
-
Yang Z, Keel SB, Shimamura A, Liu L, Gerds AT, Li HY, et al. Delayed globin synthesis leads to excess heme and the macrocytic anemia of Diamond Blackfan anemia and del(5q) myelodysplastic syndrome. Sci Transl Med. 2016;8:338ra367.
-
Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319:825–8.
Google Scholar
-
Rey MA, Duffy SP, Brown JK, Kennedy JA, Dick JE, Dror Y, et al. Enhanced alternative splicing of the FLVCR1 gene in Diamond Blackfan anemia disrupts FLVCR1 expression and function that are critical for erythropoiesis. Haematologica. 2008;93:1617–26.
Google Scholar
-
Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–52.
Google Scholar
-
Gastou M, Rio S, Dussiot M, Karboul N, Moniz H, Leblanc T, et al. The severe phenotype of Diamond-Blackfan anemia is modulated by heat shock protein 70. Blood Adv. 2017;1:1959–76.
Google Scholar
-
Das G, Shravage BV, Baehrecke EH. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol. 2012;4:a008813.
-
Doulatov S, Vo LT, Macari ER, Wahlster L, Kinney MA, Taylor AM, et al. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med. 2017;9:eaah5645.
-
Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–9.
Google Scholar
-
Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069–79.
Google Scholar
-
Stark AR, Carlo WA, Tyson JE, Papile LA, Wright LL, Shankaran S, et al. Adverse effects of early dexamethasone treatment in extremely-low-birth-weight infants. National Institute of Child Health and Human Development Neonatal Research Network. N Engl J Med. 2001;344:95–101.
Google Scholar
-
Yeh TF, Lin YJ, Huang CC, Chen YJ, Lin CH, Lin HC, et al. Early dexamethasone therapy in preterm infants: a follow-up study. Pediatrics. 1998;101:E7.
Google Scholar
-
Maceckova Z, Kubickova A, De Sanctis JB, Hajduch M. Effect of glucocorticosteroids in Diamond-Blackfan Anaemia: maybe not as elusive as it seems. Int J Mol Sci. 2022;23:1886.
-
Ashley RJ, Yan H, Wang N, Hale J, Dulmovits BM, Papoin J, et al. Steroid resistance in Diamond Blackfan anemia associates with p57Kip2 dysregulation in erythroid progenitors. J Clin Invest. 2020;130:2097–110.
Google Scholar
-
Lipton JM, Atsidaftos E, Zyskind I, Vlachos A. Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: an update from the Diamond Blackfan Anemia Registry. Pediatr Blood Cancer. 2006;46:558–64.
Google Scholar
-
Berdoukas V, Nord A, Carson S, Puliyel M, Hofstra T, Wood J, et al. Tissue iron evaluation in chronically transfused children shows significant levels of iron loading at a very young age. Am J Hematol. 2013;88:E283–85.
Google Scholar
-
Marsella M, Borgna-Pignatti C. Transfusional iron overload and iron chelation therapy in thalassemia major and sickle cell disease. Hematol Oncol Clin North Am. 2014;28:703–27.
Google Scholar
-
Peffault de Latour R, Peters C, Gibson B, Strahm B, Lankester A, de Heredia CD, et al. Recommendations on hematopoietic stem cell transplantation for inherited bone marrow failure syndromes. Bone Marrow Transpl. 2015;50:1168–72.
Google Scholar
-
Diaz-de-Heredia C, Bresters D, Faulkner L, Yesilipek A, Strahm B, Miano M, et al. Recommendations on hematopoietic stem cell transplantation for patients with Diamond-Blackfan anemia. On behalf of the Pediatric Diseases and Severe Aplastic Anemia Working Parties of the EBMT. Bone Marrow Transpl. 2021;56:2956–63.
-
Miano M, Eikema DJ, de la Fuente J, Bosman P, Ghavamzadeh A, Smiers F, et al. Stem cell transplantation for Diamond-Blackfan anemia. A Retrospective Study on Behalf of the Severe Aplastic Anemia Working Party of the European Blood and Marrow Transplantation Group (EBMT). Transpl Cell Ther. 2021;27:274.e1–274.e5.
-
Strahm B, Loewecke F, Niemeyer CM, Albert M, Ansari M, Bader P, et al. Favorable outcomes of hematopoietic stem cell transplantation in children and adolescents with Diamond-Blackfan anemia. Blood Adv. 2020;4:1760–9.
Google Scholar
-
Fagioli F, Quarello P, Zecca M, Lanino E, Corti P, Favre C, et al. Haematopoietic stem cell transplantation for Diamond Blackfan anaemia: a report from the Italian Association of Paediatric Haematology and Oncology Registry. Br J Haematol. 2014;165:673–81.
Google Scholar
-
Mamcarz E, Zhou S, Lockey T, Abdelsamed H, Cross SJ, Kang G, et al. Lentiviral gene therapy combined with low-dose busulfan in infants with SCID-X1. N Engl J Med. 2019;380:1525–34.
Google Scholar
-
Bernardo ME, Aiuti A. The role of conditioning in hematopoietic stem-cell gene therapy. Hum Gene Ther. 2016;27:741–8.
Google Scholar
-
Dahl M, Warsi S, Liu Y, Debnath S, Billing M, Siva K, et al. Bone marrow transplantation without myeloablative conditioning in a mouse model for Diamond-Blackfan anemia corrects the disease phenotype. Exp Hematol. 2021;99:44–53.e42.
Google Scholar
-
Czechowicz A, Kraft D, Weissman IL, Bhattacharya D. Efficient transplantation via antibody-based clearance of hematopoietic stem cell niches. Science. 2007;318:1296–9.
Google Scholar
-
George BM, Kao KS, Kwon HS, Velasco BJ, Poyser J, Chen A, et al. Antibody conditioning enables MHC-mismatched hematopoietic stem cell transplants and organ graft tolerance. Cell Stem Cell. 2019;25:185–92.e183.
Google Scholar
-
Dietz AC, Savage SA, Vlachos A, Mehta PA, Bresters D, Tolar J, et al. Late effects screening guidelines after hematopoietic cell transplantation for inherited bone marrow failure syndromes: Consensus Statement From the Second Pediatric Blood and Marrow Transplant Consortium International Conference on Late Effects After Pediatric HCT. Biol Blood Marrow Transpl. 2017;23:1422–8.
-
Cavazzana M, Bushman FD, Miccio A, Andre-Schmutz I, Six E. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat Rev Drug Discov. 2019;18:447–62.
Google Scholar
-
Tucci F, Galimberti S, Naldini L, Valsecchi MG, Aiuti A. A systematic review and meta-analysis of gene therapy with hematopoietic stem and progenitor cells for monogenic disorders. Nat Commun. 2022;13:1315.
Google Scholar
-
Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363:355–64.
Google Scholar
-
Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151.
Google Scholar
-
Thompson AA, Walters MC, Kwiatkowski J, Rasko JEJ, Ribeil JA, Hongeng S, et al. Gene therapy in patients with transfusion-dependent beta-thalassemia. N Engl J Med. 2018;378:1479–93.
Google Scholar
-
Ribeil JA, Hacein-Bey-Abina S, Payen E, Magnani A, Semeraro M, Magrin E, et al. Gene therapy in a patient with sickle cell disease. N Engl J Med. 2017;376:848–55.
Google Scholar
-
Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158.
Google Scholar
-
Sessa M, Lorioli L, Fumagalli F, Acquati S, Redaelli D, Baldoli C, et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet. 2016;388:476–87.
Google Scholar
-
Liu Y, Dahl M, Debnath S, Rothe M, Smith EM, Grahn THM, et al. Successful gene therapy of Diamond-Blackfan anemia in a mouse model and human CD34+ cord blood hematopoietic stem cells using a clinically applicable lentiviral vector. Haematologica. 2022;107:446–56.
-
SV Bhoopalan, J Yen, T Mayuranathan, Y Yao, K Mayberry, S Zhou, et al. A novel RPS19-edited hematopoietic stem cell model of Diamond-Blackfan anemia for development of lentiviral vector gene therapy. BLood. 2021;138:859.
-
Richard A, Voit XL, Cohen B, Armant M, Kamal E, Mei-Mei Huang W, et al. Regulated expression of GATA1 as a gene therapy cure for Diamond-Blackfan anemia. Blood. 2022;140:986–7.
-
Newby GA, Yen JS, Woodard KJ, Mayuranathan T, Lazzarotto CR, Li Y, et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 2021;595:295–302.
Google Scholar
-
Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, et al. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia. N Engl J Med. 2021;384:252–60.
Google Scholar
-
Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–24.
Google Scholar
-
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–44.
Google Scholar
-
Schiroli G, Conti A, Ferrari S, Della Volpe L, Jacob A, Albano L, et al. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell. 2019;24:551–65.e558.
Google Scholar
-
Ferrari S, Jacob A, Cesana D, Laugel M, Beretta S, Varesi A, et al. Choice of template delivery mitigates the genotoxic risk and adverse impact of editing in human hematopoietic stem cells. Cell Stem Cell. 2022;29:1428–44.e1429.
Google Scholar
-
Li C, Georgakopoulou A, Newby GA, Chen PJ, Everette KA, Paschoudi K, et al. In vivo HSC prime editing rescues sickle cell disease in a mouse model. Blood. 2023;141:2085–99.
Google Scholar
-
Raguram A, Banskota S, Liu DR. Therapeutic in vivo delivery of gene editing agents. Cell. 2022;185:2806–27.
Google Scholar
Acknowledgements
This work was supported by a Hemato-Linne grant from the Swedish Research Council Linnaeus, project grants from Swedish Research Council (to SK), the Swedish Cancer Society and the Swedish Children’s Cancer Society (to SK), the Tobias Prize awarded by the Royal Swedish Academy of Sciences financed by the Tobias Foundation, a clinical research grant from Lund University Hospital (to SK), European Union project grants STEMEXPAND and PERSIST, grant from The Royal Physiographic Society of Lund, Sweden (to YL), and grant from Stiftelsen Lars Hiertas Minne (to YL).
Funding
Open access funding provided by Lund University.
Author information
Authors and Affiliations
Contributions
Contribution: YL and SK wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Liu, Y., Karlsson, S. Perspectives of current understanding and therapeutics of Diamond-Blackan anemia.
Leukemia (2023). https://doi.org/10.1038/s41375-023-02082-w
-
Received: 18 August 2023
-
Revised: 20 October 2023
-
Accepted: 06 November 2023
-
Published: 16 November 2023
-
DOI: https://doi.org/10.1038/s41375-023-02082-w