Infection
Molecular occurrence and genetic diversity of Ehrlichia canis in naturally infected dogs from Thailand
Abstract
Canine monocytic ehrlichiosis is cause by Ehrlichia canis resulting in hematologic disorders and severe clinical signs. The aim of this study was to scrutinize the molecular detection and genetic diversity of E. canis based on the trp36 gene in dogs from Thailand’s northern and central regions. A total of 120 dogs blood samples were amplified for trp36 gene of E. canis using the polymerase chain reaction (PCR). Forty-seven out of 120 dog blood samples (39.16%, 47/120) were positive for E. canis the trp36 DNA with 790 bp of PCR amplicon size. The factor significantly associated with E. canis infection is animal housing status (p < 0.05). Sequence and phylogenetic analysis showed that E. canis trp36 gene of Thailand isolates was clustered into 1st clade with similarity ranging from 95.65 to 100% together with the US genogroup. The 14 haplotypes of the trp36 gene shown in TCS network exhibited that haplotype #1–4 was found in Thailand. The entropy analysis of the trp36 gene illustrated 751 polymorphic sites and 271 entropy peaks of nucleic and amino acid sequences, respectively. Hence, these findings are crucial for better understanding the epidemiology of Ehrlichia infection and could be helpful for implementing control measures in Thailand.
Introduction
Ehrlichia canis, an obligately intracellular bacterium transmitted by ticks that affects dogs, causes canine monocytic ehrlichiosis (CME). CME is prevalent in the tropics, particularly in Thailand and Southeast Asia1,2,3,4. E. canis can affect the dog’s monocytes and macrophages, resulting in hematologic disorders and clinical signs like fever, depressive symptoms, anorexia, weight loss, hemorrhage, epistaxis, anemia, thrombocytopenia and even death5. The occurrence of CME in dogs has been described in some Thai provinces, including Chiang Mai, Mahasarakham, Buriram, Kalasin, Nakhon Pathom, Songkha and Khonkaen, and can reach up to 36.73 percent, according to results of microscopic examination and PCR assay6,7,8,9,10,11,12.
The microscopic examination of E. canis in Giemsa-stained blood used to diagnose CME has a low sensitivity when parasitemia is low9, 13. Serological tests are an alternative method of detection that veterinarians more often use in conjunction with the rapid tests which are commercially available. However, it takes a few weeks for antibodies to occur. When diagnosing infections, particularly in laboratories, the molecular method by polymerase chain reaction (PCR) is reliable and frequently used. It provides high sensitivity and specificity in cases of low parasitemia or early stages of infection in domesticated animals13, 14. In E. canis, the tandem repeat protein 36 (TRP36) is the immunodominant protein which has been involved with host–pathogen interactions, e.g., adhesion, internalization, actin nucleation and immune evasion15,16,17. TRP36 protein is encoded by the trp36 gene containing a 5′ end pre-repeat, a tandem repeat and a 3′ end post-tandem repeat regions15, 18. Based on TR sequences, trp36 gene of E. canis strains can be divided into four genogroups including United States (US), Taiwan (TWN), Brazil (BR) and Costa Rica (CR)19,20,21. Additionally, novel TR sequences of E. canis infection were identified in humans from Costa Rica21, 22. Notably, the trp36 gene exhibited significant variability, rendering it a promising candidate for gene utilization in genetic diversity assessment and clustering6. Little is known about E. canis’s genetic diversity in Thailand1, 2, 7, 8. Therefore, the aim of this study was to scrutinize the molecular detection and genetic diversity of E. canis based on the trp36 gene in dogs from Thailand’s northern and central regions. A bioinformatics sequence analysis was also used to provide more information on the genetic profile of E. canis populations in Thailand in comparison to those found in other nations around the world.
Results
Occurrence of E. canis infection and risk factor analysis
Forty-seven out of 120 samples (39.16%) were positive for E. canis trp36 gene detected by PCR. The size of PCR product of E. canis trp36 Thailand sequence was 790 bp. Seven DNA sequences were deposited in GenBank, and accession numbers are provided in Table 1. The results of the univariate analyses regarding the overall E. canis infection detected by PCR in association with sex, age, tick infestation and animal housing status are shown in Table 2. The results showed that only animal housing status factor showed higher risk of E. canis infection in free roaming group than the dog living in owner house with statistically significant association (X2 = 11.831, p = 0.00058), while the remaining three factors exhibited no statistically significant association as shown in Table 2.
Sequence analysis of E. canis trp36 gene
E. canis trp36 sequences was divided into three regions: pre-tandem (427 bp), tandem (27 bp repeat units) and post-tandem repeat regions (none of trp36 Thailand sequence contained this region due to short sequence amplification). All sequences can be divided into four genogroups including the United States (US), Costa Rica (CR), Brazil (BR) and Taiwan (TWN) (Fig. 1).
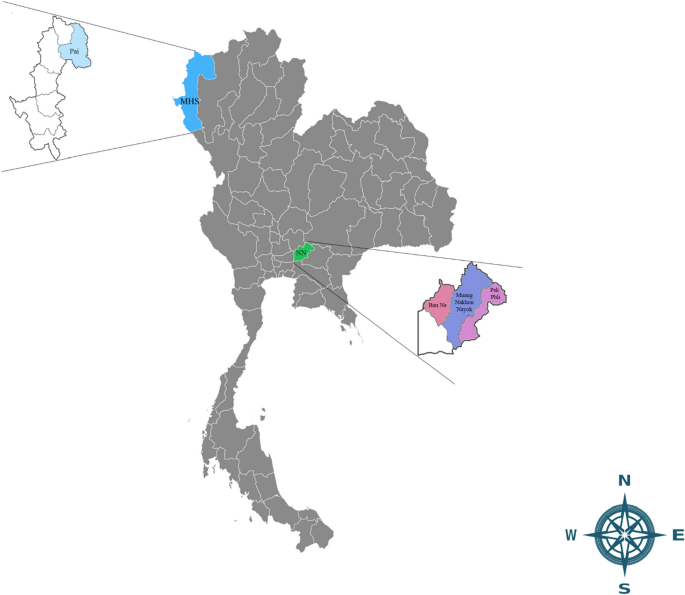
Geographical location of Mae Hong Son and Nakhon Nayok provinces where canine blood samples were collected. Legends indicate the detection of E. canis trp36 gene Thailand sequences identified in dogs from Pai district in Mae Hong Son (MHS) province and Ban Na, Muang Nakhon Nayok and Pak Phli districts in Nakhon Nayok (NN) province.
Phylogenetic and similarity analysis of E. canis and trp36 gene sequences
Seven sequences of E. canis trp36 gene obtained in this study were aligned with 19 other sequences taken from the GenBank including sequences from USA, Cameroon, Brazil, Mexico, Taiwan and Colombia. The phylogenetic tree of the trp36 gene was classified as 4 clades (designated as clade 1–4). Our Thailand sequences detected in this work were positioned in 1st clade close to the US genogroup, while clades 2, 3 and 4 were consisted of the sequence from Columbia, Brazil and Taiwan (Fig. 2). The total similarity of Thailand sequences was 95.65–100% (1st clade), while that of the sequences within each clade was 88.58–100% (1st clade, US genogroup) and 97.81–100% (2nd clade, Costa Rica genogroup), 91.72–100% (3rd clade, Brazil genogroup) and 87.25–100% (4th clade, Taiwan genogroup) as shown in Table 3. Additionally, the nucleic acid substitution rate in trp36 gene sequences among E. canis was analyzed by the Tamura and Nei mode as shown in Table 4.

A maximum likelihood phylogenetic tree relationship of E. canis trp36 gene sequences in this study (boldface) and those obtained from GenBank database. The numbers on each node correspond to the bootstrap analysis of 1000 replicates. The GenBank accession numbers of the sequences used in the phylogenetic trees are also demonstrated. A sequence of Ehrlichia chaffeensis gp47 gene is used as an outgroup. The scale measures the number of substitution per site.
Haplotype diversity analysis
The haplotype networks of E. canis trp36 gene sequences were constructed from a TCS network tool (Fig. 3). The 14 haplotypes of this gene shown in TCS network exhibited that haplotype #1–4 was found in Thailand, and the rest of the haplotypes were detected in other countries including USA, Cameroon, Brazil, Mexico, Taiwan and Colombia (Fig. 3 and Table 5).

A haplotypeTCS network based on the E. canis trp36 gene sequence isolated from Thailand and worldwide. Small traits between a haplotype and another represent mutational occurrence. The black circles are the intermediated traits caused by the single nucleotide polymorphism (SNP).
Entropy analysis
The entropy analysis of nucleotides revealed that the post-tandem region of trp36 sequences showed 751 polymorphic sites with entropy values ranged between 0.18491 and 1.46376 (Fig. 4A). Entropy analysis of amino acid sequences was conducted using the TRP36 amino acid sequence alignments. The charts exhibited 271 high entropy peaks for the TRP36 value ranging from 0.18491 to 1.75496 (Fig. 4B).

Entropy H(x) analysis of E. canis trp36 sequence. Entropy plot of multiple nucleic acid sequence alignment of trp36 genes (A). Entropy plot of multiple amino acid sequence alignment of trp36 gene (B). The red peaks refer to high variation at each position of the nucleic (A) and amino (B) acid sequences.
Discussion
In Thailand, canine monocytic ehrlichiosis (CME) caused by E. canis is a serious tick-borne disease causing severe clinical infection in dogs resulting in death1,2,3,4,5,6,7,8,9,10, 12,13,14. Some dogs show healthy appearance, but E. canis infection can be detected by PCR screening due to early phase of infection and low parasitemia level9, 13. TRP36 protein of E. canis encoded by trp36 gene can elicit in the earliest acute-phase antibody response and involves in host–pathogen interaction23. This study is the first report that revealed the infection rate, molecular characteristics and genetic diversity of E. canis in dog blood samples in Mae Hong Son and Nakhon Nayok provinces in Thailand. The molecular detection exhibited that of the dogs sampled, 39.16% (47/120) was positive for E. canis trp36 gene. The occurrence of E. canis in this study also agrees with previous reports in Thailand; for instance, 33% in Bangkok24, 36% in Chiang Mai, Nonthaburi and Chonburi provinces2 and 36.1% in Chiang Mai provinces6 By contrast, in Colombia, E. canis was found in 11.67% of sampled dogs25. The results of univariate analyses indicated that sex and age were not significant to the E. canis infection and our results were in line with previous reports of Tazawa et al.13 and Mitpasa et al.12. For tick infestation factor, the non-significant p-value (p = 0.219) indicates that there is no statistically significant difference in the frequency of E. canis between dogs parasitized by ticks and those without ticks. Most of dogs in this study appear subclinical infection that were recruited for neutralization from different areas. In previous study, Paulino et al.26 who revealed that climate change of study area can affect biological growth of Rhipicephalus sanguineus which are the vector of E. canis26. R. sanguineus has a life cycle with three-host stages and seeks a new host for a blood meal after each of its three molts, but the pathogens have already transmitted to the infected host. Additionally, dogs living in the shelter or free roaming have higher risk for E. canis infection than dogs living with owner significantly (p = 0.00058) which is consistent with other studies reported by Mitpasa et al.12 and Navarrete et al.27.
Although the genetic diversity of E. canis strains based on the trp36 gene has been characterized to 4 genogroups in several countries19, 27. There is very little information regarding the genetic diversity and phylogenetic analysis of E. canis trp36 gene in Thailand so far. The phylogeny analysis of E. canis trp36 gene Thailand isolates showed totally only one clade with other strains. Bootstrap values in the phylogenetic tree in this study were 78-100% of bootstrap values, which are in line with a majority-rule consensus tree of 1000 replicates for each alignment28, 29. The results showed that the genetic diversity and phylogenetic proximity of the E. canis trp36 gene to the US sequences (US genogroup) were evident from the conserved nucleotide sequence TACTGAAGATTCTGTTTCTGCTCCAGC, which translated to the amino acid sequence TEDSVSAP in the tandem repeat region. This classification grouped Thai samples together with other sequences from the US genogroup in the same clade, showing a similarity range of 88.58–100%. Comparatively, the US genogroup displayed less diversity within the group when compared to the other genogroups in the TCS network. The main differing conserved region were classified by the tandem repeat region of the E. canis trp36 gene. This finding was similar to previous study in Nonthaburi, Chonburi and Chiang Mai provinces of Thailand reported by Poolsawat et al.2 and Nambooppha et al.6. This finding indicated the phylogenetic proximity of E. canis trp36 gene circulating in both different countries and Thailand. Our finding is similar to the previous studies reported by da Costa et al.30 and Kaewmonkol et al.24.
The trp36 gene distinguishes itself as an appropriate genotyping marker for E. canis strains due to its alleles encoding distinct TR amino acid sequences of TRP36. Its utility extends to the assessment of genetic diversity among E. canis isolates, revealing pronounced variations in TR sequences and/or TR numbers across diverse geographic regions19, 20, 31. The most preserved TR in E. canis strains worldwide is TEDSVSAPA from the US genotype, and a similar preservation is observed in Taiwan genogroups with different N-terminal pre-TR regions17, 19. A novel Brazilian genotype has been reported with a different tandem repeat sequence (ASVVPEAE) in dog samples in Brazil. However, some dog samples in Brazil exhibit a pre-TR region similar to US genogroups17, 20. A novel genotype consisting of one TR with the sequence EASVVPAAEAPQPAQQTEDEFFSDGIEA was reported in the Costa Rica (Cr) genogroup21. Moreover, TR sequences of EASVVPAAEAPQPAQQTEDEFFSDGIEA and EASVVPAAEAPQPAQQTEDEFFSDGIE amino acid sequences were identified in humans from Costa Rica22. In many studies, some isolates in the same country were classified into different genogroups depending on their sequences. For instance, in the study of Turkish isolates of E. canis, it was reported that the Turkish isolate sequences were segregated into four distinct genogroups: US genogroups I and II, Brazilian genogroup, and Costa Rica-Turkey genogroup. Seven E. canis Turkish isolates and E. canis-human Costa Rica were placed in a new genogroup designated in this study as Costa Rica-Turkey genogroup22.
In this study, our Thailand samples were genetically conserved and closed to the US genogroup sequences as shown in TCS network and shared genetic traits with other sequences as retrieved previously worldwide. The Taiwan and Brazil genogroups contain single-nucleotide polymorphism (SNP) trait different from Thailand sequences related to the different of nucleotide base and translated amino acid in tandem repeat and post-tandem repeat regions of the trp36 gene. The high SNP variations, which are linked to a high number of nucleotide and amino acid variables, are shown by the high entropy values and polymorphic sites. The lower entropy values reveal that each sequence contains few SNP variants32.
The genetic diversity observed in the trp36 gene, particularly in the tandem repeat region, has revealed a potential novel target for organism genotyping. This study’s findings contribute to our understanding of E. canis’ genetic diversity and highlight the importance of further research to analyze genetic variations in E. canis strains worldwide. TRP36 protein, encoded by the trp36 gene (DQ146154 in GenBank)18, exhibits distinct expression patterns within the dense-cored morphological variant of Ehrlichia. In this form, the protein is both exposed on the cell surface and secreted15. TRP36 protein of E. canis represents an immunodominant protein, playing a significant role in host-pathogen interactions and triggering the earliest acute-phase antibody response during the disease progression15. Its recognition as a surface protein early in the infection process makes TRP36 a promising candidate for diagnostic tools and vaccine development15, 23.
Conclusions
This study is the first report regarding a molecular occurrence and genetic diversity of E. canis in canine samples from Thailand’s Mae Hong Son and Nakhon Nayok provinces. Our results revealed that the diversity of E. canis trp36 gene is genetically conserved in Thailand and worldwide. These results may help to clarify the molecular phylogeny and diversity of the trp36 genes of E. canis Thailand strains. Hence, our finding may be useful in immunodiagnostic tools and vaccination for CME.
Methods
Sample population
This study was conducted during October 2022 to March 2023. A total of 120 blood samples from canine shelters in the north (17 dogs from Pai district; Mae Hong Son province, 19° 22′ 51.222″ N latitude, 98° 26′ 40.1064″ E longitude) and central (103 dogs from Ban Na, Muang Nakhon Nayok, Pak Phli district; Nakhon Nayok province, 14° 13′ 7.608″ N latitude, 101° 18′ 24.84″ E longtitude) regions of Thailand, were used in this study (Fig. 1). The sample sizes were calculated using the formula based on the equation, n = t2 × p (1 − p)/m2, inserting the following values: the prevalence (p) of E. canis infection among dogs in Thailand, a 95% confidence level (t) and 5% margin of error (m)1, 13.
Collection of blood samples
Approximately three ml of whole blood samples were obtained from the cephalic or lateral saphenous veins of each animal, collected in EDTA-tubes (BD Vacutainer®, USA) and kept at − 20 °C. Additionally, licensed veterinarians carried out the processes of animal restraint and blood sample collection.
DNA extraction and PCR amplification of the trp36 gene of E. canis
Genomic DNA of E. canis was extracted from dogs’ blood samples using a DNA Extraction Kit (OMEGA, bio-tex, USA) according to the protocol of Junsiri et al.33,34,35, Poolsawat et al.1, 2 and Watthanadirek et al.36 with some modifications. Briefly, the DNA sample was eluted in 30 µl MiliQ water and concentration of purified DNA sample was defined with NanoDrop™ 2000 Spectrophotometers (Thermo Scientific™, USA) at the 260/280 and 260/230 ratios. Finally, the aliquots were stored at − 20 °C until further use. The trp36 gene was amplified by single PCR using the specific primers: TRP36F 5′-ATGCTACTTTTACTAATGGGTTATTGT-3′ and TRP36R 5′-GTACAACATGTTAAGAATATCAG-3′24 according to the protocol of Poolsawat et al.2. For PCR reaction, 50 ng of purified DNA template was added in a total volume of 25 μl of reaction mixture containing 0.2 μM of each primer, 200 μM of each deoxynucleoside triphosphate (dNTPs), 1 × phusion HF buffer, nuclease free water and 0.5 U Phusion® High-Fidelity DNA Polymerase (NEW ENGLAND BioLabs®Inc, USA). The thermocycling protocol for the trp36 gene was carried out with the conditions: 98 °C for 3 min followed by 35 cycles at 98 °C for 60 s, 56 °C for 60 s, 72 °C for 90 s, and 72 °C for 5 min. The PCR amplicon was stained with FluoroStain™ DNA Fluorescent Staining Dye (SMOBIO®, Taiwan). PCR products were visualized with gel electrophoresis using 1% agarose gel under UV illumination and photographed. A 100 bp DNA Ladder M (SMOBIO®, Taiwan) was used as a standard for defining the molecular mass of PCR products.
Molecular cloning and sequencing of E. canis trp36 gene
The purified PCR product was cloned into the pGEM®-T Easy vector (Promega, USA). The ligation product was transformed into the Escherichia coli strain DH5-alpha cells (Invitrogen, USA). Then, the transformed E. coli cells were cultured on the Luria Bertani (LB) medium agar plate supplemented with ampicillin (100 μg/ml) and X-GAL (20 mg/ml). After incubation at 37 °C overnight, the white colonies were selected and grown in LB medium containing ampicillin for overnight. Finally, the recombinant plasmid (pGEM®-T-trp36) was extracted from the competent cell using the Presto™ Mini Plasmid Kit (Geneaid, Taiwan) following the manufacturer’s instructions, and analyzed for accurate sized inserts by agarose gel electrophoresis. The presence of trp36 insert was confirmed by Sanger sequencing. All sequences were analyzed by BLAST (The National Center for Biotechnology Information, NCBI, http://www.ncbi.nlm.nih.gov/ BLAST), and deposited in the GenBank database.
Phylogenetic tree analysis
The E. canis trp36 gene sequences were aligned with Muscle algorithm, and genetic inference was carried out with phylogenetic tree which was reconstructed using the maximum likelihood (ML) as implemented in the MEGA software v.7.0.2637. Bootstrap analysis with 1000 repetitions was used to assess the reliability of the branching pattern of the ML trees38. The evolutionary distance was analyzed by the Hasegawa–Kishino–Yano model39. The similarity of nucleotide sequences was evaluated by a sequence identity matrix in Bioedit software v.7.0.5.340.
Analysis of haplotype diversity
The sequences alignment of E. canis trp36 gene was employed to evaluate the nucleotide diversity (π), diversity of haplotypes (Dh), number of haplotypes (h), and the average number of nucleotide differences (K), using the DnaSP v.6.12.0341. All sequences were subjected to the popART program42 to construct the TCS Network43.
Entropy analysis
Entropy estimation was employed to ascertain the variability of the nucleotide and amino acid sequences of E. canis. The E. canis trp36 nucleotide sequences were translated into amino acid sequences, aligned and analyzed by the entropy (H (x)) plot using Bioedit software v.7.0.5.340.
Statistical analysis
The demographic factors and the overall infection status were analyzed in relation to the infection using Pearson’s Chi-squared test. The relationship between risk factors and occurence was analyzed using the logistic regression test with p-value < 0.05 in SPSS software v. 22.0 (IBM Corp., NY, USA) (IBM Corp., 2013)44.
Ethics statement
All experimental procedures on animals were approved by the Animal Care and Use Committee (IMBMU-ACUC), Institute of Molecular Biosciences, Mahidol University, Thailand. All biological samples were collected with authorized consent form from the canine shelter and hospital. In addition, all methods were performed in accordance with the relevant guidelines and regulations.
Data availability
The datasets generated and/or analysed during the current study are available in the GenBank database, and the accession number are OP748407, OP748408, OP748409, OP748418, OP748419, OP748420, and OP748421. The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
-
Poolsawat, N. et al. Molecular discrimination and genetic diversity of three common tick-borne pathogens in dogs in Thailand. Parasitology 149, 65–75. https://doi.org/10.1017/S0031182021001566 (2022).
Google Scholar
-
Poolsawat, N. et al. Ehrlichia canis: Molecular characterization and genetic diversity based on the p28 and trp36 genes. Res. Vet. Sci. 155, 88–102. https://doi.org/10.1016/j.rvsc.2022.11.013 (2023).
Google Scholar
-
Maekawa, N. et al. Molecular detection and phylogenetic analysis of Ehrlichia canis in a Philippine dog. Ticks Tick Borne Dis. 9, 266–269. https://doi.org/10.1016/j.ttbdis.2017.09.012 (2018).
Google Scholar
-
Immelman, A. & Button, C. Ehrlichia canis infection (Tropical canine pancytopaenia or canine rickettsiosis). J. S. Afr. Vet. Assoc. 44, 241–245 (1973).
Google Scholar
-
Saito, T. B. & Walker, D. H. Ehrlichioses: An important one health opportunity. Vet. Sci. 3, 20. https://doi.org/10.3390/vetsci3030020 (2016).
Google Scholar
-
Nambooppha, B. et al. Two different genogroups of Ehrlichia canis from dogs in Thailand using immunodominant protein genes. Infect. Genet. Evol. 63, 116–125. https://doi.org/10.1016/j.meegid.2018.05.027 (2018).
Google Scholar
-
Piratae, S., Pimpjong, K., Vaisusuk, K. & Chatan, W. Molecular detection of Ehrlichia canis, Hepatozoon canis and Babesia canis vogeli in stray dogs in Mahasarakham province Thailand. Ann. Parasitol. 61, 183–187. https://doi.org/10.17420/ap6103.05 (2015).
Google Scholar
-
Piratae, S., Senawong, P., Chalermchat, P., Harnarsa, W. & Sae-Chue, B. Molecular evidence of Ehrlichia canis and Anaplasma platys and the association of infections with hematological responses in naturally infected dogs in Kalasin, Thailand. Vet. World 12, 131–135. https://doi.org/10.14202/vetworld.2019.131-135 (2019).
Google Scholar
-
Rucksaken, R., Maneeruttanarungroj, C., Maswanna, T., Sussadee, M. & Kanbutra, P. Comparison of conventional polymerase chain reaction and routine blood smear for the detection of Babesia canis, Hepatozoon canis, Ehrlichia canis, and Anaplasma platys in Buriram Province, Thailand. Vet. World 12, 700–705. https://doi.org/10.14202/vetworld.2019.700-705 (2019).
Google Scholar
-
Liu, M. et al. Molecular survey of canine vector-borne diseases in stray dogs in Thailand. Parasitol. Int. 65, 357–361. https://doi.org/10.1016/j.parint.2016.04.011 (2016).
Google Scholar
-
Harrus, S. & Waner, T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): An overview. Vet. J. 187, 292–296. https://doi.org/10.1016/j.tvjl.2010.02.001h (2011).
Google Scholar
-
Mitpasa, T. et al. First report on molecular characteristics and risk factor analysis of Ehrlichia canis in dogs in Khon Kaen, Thailand. Vet. World 15, 232–238. https://doi.org/10.14202/vetworld.2022.232-238 (2022).
Google Scholar
-
Tazawa, K. et al. First study on molecular detection of three major canine tick-borne pathogens in subclinically infected dogs in Chiang Mai, Thailand. Vet. World 15, 1121–1128. https://doi.org/10.14202/vetworld.2022.1121-1128 (2022).
Google Scholar
-
Aktas, M. et al. Molecular detection of tick-borne rickettsial and protozoan pathogens in domestic dogs from Turkey. Parasit. Vectors 8, 157. https://doi.org/10.1186/s13071-015-0763-z (2015).
Google Scholar
-
Doyle, C. K., Nethery, K. A., Popov, V. L. & McBride, J. W. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect Immun. 74, 711–720. https://doi.org/10.1128/IAI.74.1.711-720.2006 (2006).
Google Scholar
-
McBride, J. W. & Walker, D. H. Molecular and cellular pathobiology of Ehrlichia infection: Targets for new therapeutics and immunomodulation strategies. Expert Rev. Mol. Med. 13, e3. https://doi.org/10.1017/S1462399410001730 (2011).
Google Scholar
-
Aguiar, D. M. et al. Genetic diversity of Ehrlichia canis in Brazil. Vet. Microbiol. 164, 315–321. https://doi.org/10.1016/j.vetmic.2013.02.015 (2013).
Google Scholar
-
Doyle, C. K. et al. Molecular characterization of E. canis gp36 and E chaffeensis gp47 tandem repeats among isolates from different geographic locations. Ann. N. Y. Acad. Sci. 1063, 433–435. https://doi.org/10.1196/annals.1355.079 (2005).
Google Scholar
-
Hsieh, Y. C., Lee, C. C., Tsang, C. L. & Chung, Y. T. Detection and characterization of four novel genotypes of Ehrlichia canis from dogs. Vet. Microbiol. 146, 70–75. https://doi.org/10.1016/j.vetmic.2010.04.013 (2010).
Google Scholar
-
Aguiar, D. M. & Melo, A. L. Divergence of the TRP36 protein (gp36) in Ehrlichia canis strains found in Brazil. Ticks Tick Borne Dis. 6, 103–105. https://doi.org/10.1016/j.ttbdis.2014.10.003 (2015).
Google Scholar
-
Aktas, M. & Özübek, S. Genetic diversity of Ehrlichia canis in dogs from Turkey inferred by TRP36 sequence analysis and phylogeny. Comp. Immunol. Microbiol. Infect. Dis. 64, 20–24. https://doi.org/10.1016/j.cimid.2019.02.003 (2019).
Google Scholar
-
Bouza-Mora, L. et al. Novel genotype of Ehrlichia canis detected in samples of human blood bank donors in Costa Rica. Ticks Tick Borne Dis. 8, 36–40. https://doi.org/10.1016/j.ttbdis.2016.09.012 (2017).
Google Scholar
-
McBride, J. W. et al. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71, 2516–2524. https://doi.org/10.1128/IAI.71.5.2516-2524.2003 (2003).
Google Scholar
-
Kaewmongkol, S. et al. Detection of specific IgM and IgG antibodies in acute canine monocytic ehrlichiosis that recognize recombinant gp36 antigens. Heliyon 6, e04409. https://doi.org/10.1016/j.heliyon.2020.e04409 (2020).
Google Scholar
-
Arroyave, E. et al. Ehrlichia canis TRP36 diversity in naturally infected-dogs from an urban area of Colombia. Ticks Tick Borne Dis. 11, 101367. https://doi.org/10.1016/j.ttbdis.2019.101367 (2020).
Google Scholar
-
Paulino, P. G. et al. Epidemiology of Ehrlichia canis in healthy dogs from the Southeastern region of the state of Rio de Janeiro, Brazil. Prev. Vet. Med. 159, 135–142. https://doi.org/10.1016/j.prevetmed.2018.09.012 (2018).
Google Scholar
-
Navarrete, M. G. et al. Novel Ehrlichia canis genogroup in dogs with canine ehrlichiosis in Cuba. Parasit. Vectors 15, 295. https://doi.org/10.1186/s13071-022-05426-0 (2022).
Google Scholar
-
Buckley, T. R. & Cunningham, C. W. The effects of nucleotide substitution model assumptions on estimates of nonparametric bootstrap support. Mol. Biol. Evol. 19, 394–405. https://doi.org/10.1093/oxfordjournals.molbev.a004094 (2002).
Google Scholar
-
Soltis, P.S., Soltis, D.E. Applying the bootstrap in phylogeny reconstruction. Stat. Sci. 256–267 (2003).
-
da Costa, R. L. et al. Molecular characterization of Ehrlichia canis from naturally infected dogs from the state of Rio de Janeiro. Braz. J. Microbiol. 50, 1–12 (2019).
Google Scholar
-
Zhang, X. et al. Genetic and antigenic diversities of major immunoreactive proteins in globally distributed Ehrlichia canis strains. Clin. Vaccine Immunol. 15, 1080–1088. https://doi.org/10.1128/CVI.00482-07 (2008).
Google Scholar
-
Pham, T. D. GeoEntropy: A measure of complexity and similarity. Pattern Recognit. 43, 887–896 (2010).
Google Scholar
-
Junsiri, W. et al. Anaplasma marginale: Molecular discrimination, recombinant expression and characterization of major surface protein 2. Res. Vet. Sci. 152, 372–386. https://doi.org/10.1016/j.rvsc.2022.08.019 (2022).
Google Scholar
-
Junsiri, W. et al. Molecular detection and genetic diversity of Anaplasma marginale based on the major surface protein genes in Thailand. Acta Trop. 205, 105338 (2020).
Google Scholar
-
Junsiri, W. et al. Molecular characterization of Anaplasma marginale based on the msp1a and msp1b genes. Vet. Microbiol. 262, 109236. https://doi.org/10.1016/j.vetmic.2021.109236 (2021).
Google Scholar
-
Watthanadirek, A. et al. Molecular and recombinant characterization of major surface protein 5 from Anaplasma marginale. Acta Trop. 220, 105933. https://doi.org/10.1016/j.actatropica.2021.105933 (2021).
Google Scholar
-
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Google Scholar
-
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x (1985).
Google Scholar
-
Hasegawa, M., Kishino, H. & Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. https://doi.org/10.1007/BF02101694 (1985).
Google Scholar
-
Hall, T. A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98 (1999).
Google Scholar
-
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. https://doi.org/10.1093/molbev/msx248 (2017).
Google Scholar
-
Leigh, J. W. & Bryant, D. PopART: Full-feature software for haplotype network. Methods Ecol. Evol. 6, 1110–1116 (2015).
Google Scholar
-
Clement, M., Posada, D. & Crandall, K. A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. https://doi.org/10.1046/j.1365-294x.2000.01020.x (2000).
Google Scholar
-
IBM Corp. IBM SPSS Statistics for Windows, Version 22.0 (IBM Corp., Armonk, 2013).
Acknowledgements
We would like to thank Jai Dog Rescue for their technical assistance.
Funding
This work was financially supported by Mahidol University (Basic Research Fund: fiscal year 2022) and Research Grants from the Thailand Research Fund (TRF), Thailand Science Research and Innovation (TSRI), and the National Research Council of Thailand (NRCT) to Panat Anuracpreeda, as well as the Thailand Research Fund (TRF), Thailand Science Research and Innovation (TSRI), National Research Council of Thailand (NRCT) and Royal Golden Jubilee PhD (RGJ-PHD) Scholarship [grant number PHD/0055/2561] to Napassorn Poolsawat.
Author information
Authors and Affiliations
Contributions
N.P.: Conceptualization, Methodology, Validation, Investigation, Writing Original Draft, Visualization, Project administration; S.S: Resources; T.J.: Resources, Methodology; A.W.: Resources, Methodology; N.S.: Resources, Methodology; S.M.: Resources, Methodology; K.T.: Resources; Methodology; P.A.: Conceptualization, Research Design, Methodology, Validation, Investigation, Data Curation, Data Analysis, Writing—Review & Editing, Visualization, Supervision, Project administration, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Poolsawat, N., Sangchuai, S., Jaroensak, T. et al. Molecular occurrence and genetic diversity of Ehrlichia canis in naturally infected dogs from Thailand.
Sci Rep 13, 20394 (2023). https://doi.org/10.1038/s41598-023-47784-4
-
Received: 07 August 2023
-
Accepted: 18 November 2023
-
Published: 21 November 2023
-
DOI: https://doi.org/10.1038/s41598-023-47784-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.