Cancer and neoplasms
The immunome of mobilized peripheral blood stem cells is predictive of long-term outcomes and therapy-related myeloid neoplasms in patients with multiple myeloma undergoing autologous stem cell transplant
Abstract
Upfront autologous stem cell transplant (ASCT) is the standard of care for newly diagnosed multiple myeloma (MM) patients. However, relapse is ubiquitous and therapy-related myeloid neoplasms (t-MN) post-ASCT are commonly associated with poor outcomes. We hypothesized that the enrichment of abnormal myeloid progenitors and immune effector cells (IEC) in the peripheral blood stem cells (PBSCs) is associated with a higher risk of relapse and/or development of t-MN. We performed a comprehensive myeloid and lymphoid immunophenotyping on PBSCs from 54 patients with MM who underwent ASCT. Median progression-free (PFS), myeloid neoplasm-free (MNFS), and overall survival (OS) from ASCT were 49.6 months (95% CI: 39.5-Not Reached), 59.7 months (95% CI: 55–74), and 75.6 months (95% CI: 62–105), respectively. Abnormal expression of CD7 and HLA-DR on the myeloid progenitor cells was associated with an inferior PFS, MNFS, and OS. Similarly, enrichment of terminally differentiated (CD27/CD28–, CD57/KLRG1+) and exhausted (TIGIT/PD-1+) T-cells, and inhibitory NK-T like (CD159a+/CD56+) T-cells was associated with inferior PFS, MNFS, and OS post-transplant. Our observation of abnormal myeloid and IEC phenotype being present even before ASCT and maintenance therapy suggests an early predisposition to t-MN and inferior outcomes for MM, and has the potential to guide sequencing of future treatment modalities.
Introduction
Autologous stem cell transplantation (ASCT) and maintenance form the treatment backbone for most patients with multiple myeloma (MM) [1]. Along with the incorporation of novel immune-based treatment strategies, this paradigm has led to a consistent improvement in the outcomes of patients over the past two decades [2]. Despite this, a cure for MM remains elusive and most patients relapse. In addition, 2–4% of patients develop therapy-related myeloid neoplasms (t-MN) which are associated with a dismal survival, significantly contributing to post-ASCT mortality [3,4,5]. Thus, relapse and t-MN represent major barriers to improve the post-ASCT outcomes in MM. Therapy-related myeloid neoplasms are highly enriched in pathogenic variants in the tumor suppressor gene TP53 (TP53mut), which explains their aggressive course and the lack of response to the currently available therapies [6]. There is emerging concern that chimeric antigen receptor T cell therapy (CAR-T) can be complicated by the development of t-MN in upto 10–20% of patients [7, 8].
Given that ASCT is associated with significant morbidity and resource utilization, there is a substantial interest in identifying high-risk patients for relapse and t-MN before ASCT. While the focus thus far has been the neoplastic cells, recently the role of tumor immunosurveillance has emerged. A permissive immune tumor microenvironment (iTME) contributes to treatment resistance and progression of MM [9, 10]. Increased T-cell exhaustion and immune senescence have been associated with poor outcomes after standard MM therapies, ASCT and novel T cell redirecting therapies [11,12,13,14,15,16]. Immune reconstitution following autologous [17] and allogeneic SCT [18] impacts the post-transplant outcomes and immunomodulation is being pursued to improve long-term outcomes.
Similarly, TP53mut MN are characterized by a highly immunosuppressed immune milieu associated with a reduced number of immune effector cells (IEC) including cytotoxic T- and natural killer (NK)-cells, as well as the expansion of regulatory T-cells (Tregs), myeloid-derived suppressor cells (MDSC), and exhausted T-cells [19,20,21,22]. The expression of various exhaustion receptors is increased following hypomethylating agents and some patients with myelodysplastic syndromes can respond to checkpoint blockade [21, 22]. Finally, clonal expansion of terminally differentiated, immune senescent T-cells is seen in all MDS subgroups indicative of an ineffective T-cell response that fails to control disease progression [23, 24]. Peripherally mobilized blood-derived stem cell (PBSC) grafts are responsible for the reconstitution of the myeloid and immune compartments following the myeloablative doses of melphalan used in ASCT. We hypothesized that pre-existing immune abnormalities in the mobilized PBSC will lead to reconstitution of an immunosupressive iTME and contribute to early relapse and/or t-MN development. To that end, we explored the immune composition of mobilized PBSC and its relationship with subsequent MM relapse and t-MN development.
Methods
Study population
After institutional review board approval, we screened patients with active MM that were evaluated at Mayo Clinic, Rochester, MN between 01/01/2003 and 12/31/2020. Patients who underwent an ASCT and had cryopreserved PBSC products available for research were included. We enriched this cohort with patients who developed t-MN to study this group. t-MN was defined using the 2016 World Health Organization criteria [25]. The revised international staging system (R-ISS) was used to risk stratify MM patients [26]. Hematologic response to therapy and engraftment syndrome were defined per internationally accepted criteria [27, 28]. Short duration of remission for MM following ASCT was defined as <24 months if no post-ASCT maintenance was utilized or <48 months if post-ASCT maintenance was not utilized.
Mass cytometry
The antibody panels included 37 lymphoid- and myeloid-based markers each (Supplementary Table 1). For primary conjugations, purified antibodies were obtained in carrier protein-free phosphate-buffered saline and labeled using the X8 polymer MaxPAR antibody conjugation kit (Fluidigm) according to the manufacturer’s protocol. All antibodies were titrated to optimal staining concentrations using peripheral blood mononuclear cells. Antibody master mixes were prepared fresh for each experiment. All samples were processed identically. PBSC were collected after mobilization using peripheral blood leukapheresis. Using validated processes in a Current Good Manufacturing Practice (cGMP) laboratory, the cells were concentrated and mixed with a cryopreservation solution to give a final white blood cell concentration of 300 × 106 nucleated cells/ml in 10% DMSO, 10% plasma, and 30% PlasmaLyte-A (Baxter, Deerfield, IL) or Normosol-R (Hospira, Lake Forest, IL). The cells were then frozen using a controlled rate freezing process and stored in vapor phase liquid nitrogen tanks. The main products were stored in cryobags ranging from 50–100 ml in addition to 1.5 ml cryovials for additional testing as needed. Cryopreserved cells were resuscitated for mass cytometry analyses by rapid thawing and were rested in RPMI 1640 (20% fetal bovine serum) for 60 min prior to staining. Staining was performed using Fluidigm’s protocol. Briefly, 1–3 million cells were stained for viability with 5 mM cisplatin for 5 min at room temperature and quenched with cell staining medium (CSM; Fluidigm). Cells were then incubated for 10 min at room temperature with human FcR blocking reagent (Biolegend) and stained with the surface antibody cocktail for 60 min at 4 °C with gentle agitation. Finally, cells were washed twice with CSM, fixed with 1.6% paraformaldehyde, washed with CSM, and resuspended in 1:1000 solution of Iridium intercalator diluted in MaxPar Fix and Perm buffer (Fluidigm) for 20 min at room temperature. Prior to the acquisition, cells were washed twice in CSM and twice in deionized water and were then diluted to a concentration of 0.5 million cells per milliliter in water containing 10% of EQ 4 Element Beads (Fluidigm). Cells were filtered through a 35-μm membrane prior to mass cytometry acquisition. Samples were then acquired on a Helios mass cytometer.
Mass cytometry data analysis
Flow cytometry standard files were normalized and concatenated using the Fluidigm acquisition software. Flow cytometry standard files were uploaded to the OMIQ software from Dotmatics (www.omiq.ai, www.dotmatics.com), where transformation and cleaning (doublets, debris), were done as previously described [11]. To correct for between-sample technical variations (“batch effects”), we used fdaNorm [29] within the omiq.ai platform. Characteristic images of marker expression before and after normalization are shown in Supplementary Fig. 1. Clustering and visualization were performed within the omiq.ai platform using PhenoGraph [30] and UMAP [31], respectively. When clustering, 50,000 CD45+ events per file were used when using the lymphoid panel due to computational constraints. When using the myeloid panel, we manually excluded T-, B-, and NK-cells using canonical markers (CD3, CD19, and a combination of CD7 and CD56, respectively) and clustered the remaining cells and using 50,000 CD45+ events per file. Identified clusters were then exported for downstream statistical analyses. All immune subset frequencies are reported as a percent of CD45+ cells. All raw mass cytometry data are uploaded to FlowRepository (https://flowrepository.org/).
Statistical methods
Fisher’s exact test was used to compare categorical variables and Wilcoxon rank sum (2 groups) or Kruskal-Wallis test ( > 2 groups) for continuous variables. Kaplan-Meier survival analysis was used to estimate the overall survival (OS) from diagnosis of MM, and progression-free survival (PFS) and tMN-free survival (MNFS) from ASCT. The log-rank test was used to compare groups. A Cox proportional hazards model was used to perform multivariable analyses for time to event outcomes. Patients alive and without progression to MM or t-MN, were censored for PFS and MNFS analyses, respectively. Hierarchical clustering of patients according to the abundance of immune subsets was performed using Ward’s minimum variance method. Principal component analyses were performed using log transformed and scaled values of immune subset frequencies. Statistical analyses were performed using the JMP Pro statistical software version 14.1 (SAS Institute, Cary, NC). Correlation analyses were performed in R using the corrplot package. A 2-sided false discovery rate (FDR) adjusted P-value of <0.05 was considered significant when multiple comparisons were performed; otherwise, a P-value of <0.05 was considered significant.
Results
The baseline clinical characteristics of the 54 patients included in the study are shown in Table 1. Given the referral nature of our institution, patients with high-risk diseases were overrepresented. Also, given the limitations imposed by sample availability, we included patients that had PBSCs collected but did not receive lenalidomide maintenance, which became standard of care later in the study time frame. As a result, 19 patients did not receive post ASCT maintenance.
The immune landscape of mobilized PBSC
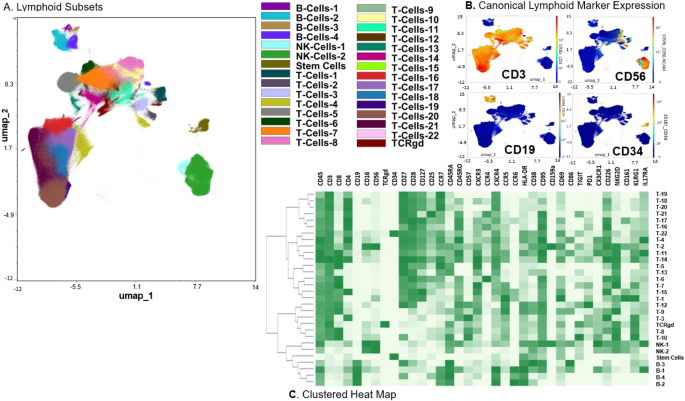
We generated 2-dimensional UMAP maps of the data generated by the 2 panels (lymphoid and myeloid) in the manually gated CD45+ cells (Supplementary Figs. 2 and 3, respectively). These analyses demonstrated that major lineages separated well in the 2-dimensional space. The most abundant cell populations were T-cells (49% of the CD45+ cells) and myeloid cells (43% of the CD45+ cells, Supplementary Fig. 4), which was not surprising considering these were mobilized cells. We then clustered all CD45+ cells using the lymphoid panel and projected the identified clusters on the respective UMAP (Fig. 1A, B). A heatmap of the marker expression of the lymphoid populations identified with the lymphoid panel is shown in Fig. 1C. To better characterize T- and NK-cell diversity, we subclustered on the manually gated respective cell subsets and excluded the markers not expressed on T- and NK-cells, respectively. Subclustering on T-cells did not increase the T-cell cluster diversity (not shown). Therefore, only the T-cell clusters that resulted after clustering with the lymphoid panel on CD45+ cells were considered for further analyses. When subclustering on NK-cells however, an additional 16 NK-cell clusters were identified and were projected on a 2-dimensional UMAP map (Fig. 2A, B). The corresponding heatmap of their marker expression is shown in Fig. 2C. We then subclustered on manually gated myeloid cells (excluding markers not expressed on myeloid cells) and identified 27 myeloid clusters which were projected on a UMAP map (Fig. 3A, B). A heatmap of their marker expression is shown in Fig. 3C.
A Phenograph Clusters projected on UMAP plot of all identified lymphoid subsets (NK, T, and B cells) and CD34+ cells using the lymphoid panel. Myeloid or lineage negative subsets were excluded for clarity. B Canonical marker expression (CD3, CD19, CD56, CD34) of major lineages. C Clustered heatmap showing the expression of all markers (lymphoid panel) expressed in identified lymphoid subsets across all files.

A Phenograph clusters projected on UMAP plot of all identified NK cell subsets after sub-clustering manually gated NK Cells and using the lymphoid panel. B Canonical NK-cell marker expression (CD16, CD56). C Clustered heatmap showing the expression of all markers expressed in NK cell subsets after sub-clustering of manually gated NK cells.

A Phenograph Clusters projected on UMAP output of all identified myeloid subsets identified by clustering all CD45+ non-T, B, or NK cells. Lineage-negative clusters not expressing canonical myeloid markers (CD11c, CD33) were excluded. B Canonical marker expression (CD33, CD11c, CD123). C Clustered heatmap showing the expression of all markers expressed in myeloid cell subsets after sub-clustering of manually gated myeloid cells.
Next, we explored the associations between the immune subsets and the dichotomized clinical outcomes of interest. To identify if specific immune subsets correlated with short or long remissions, the frequencies of the identified immune subsets between the 2 groups were compared. We found no differences at our prespecified level of significance (FDR corrected P-value < 0.05). When considering trends (non-FDR corrected P-value < 0.05), the activated (CD69/CD161/KLRG1+) memory (CD27/CD28/CD127+) CD8 T-1 cell subset was enriched in the patients with long remission compared to those with short remission (0.9% vs. 0.3%, P = 0.02). Similarly, no significant differences between the patients who subsequently developed t-MN compared to those who did not were identified. Several trends of populations of potential interest became apparent (Supplementary Table 2). NK-cell subcluster-7, characterized by CD56high, CD159a+ (NKG2A) phenotype, was enriched in the patients who subsequently developed t-MN (0.33% vs. 0.17%, P = 0.006). Enrichment of these well-described inhibitory NKG2A+ NK-cells has been noted in the bone marrow of patients with MDS and AML [32]. A “terminally mature” [33, 34] CD56/CD16/CD27dim, CCR4/CXCR3/CCR7high NK-15 subset was enriched in patients developing t-MN (0.09% vs. 0.06%, P = 0.001), and is thought to have T-cell immunosuppressive properties [35]. The T-cell subsets T-5 and T-20, two CD8+ naive subsets with bone marrow homing (CXCR4 + ) [36] and cytotoxic (NKG2D + , CD226 + , respectively) receptors were decreased in patients who later developed tMN as was the T-1 T cell subset described above. These observations imply that changes associated with decreased immunosurveillance may be evident in G-CSF mobilized PBSC products years before the development of t-MN.
We then compared patients that had been mobilized with G-CSF alone or G-CSF and plerixafor and found no differences (including those with a non-FDR corrected p-value < 0.05), suggesting that plerixafor use does not significantly influence the immune contexture of mobilized PBSC.
The structure of the immune landscape correlates with clinical outcomes
We hypothesized that the immune subsets with highly correlated frequencies were more likely to be co-regulated. A correlation matrix and a list of all identified significant (P < 0.05) correlations sorted by their R values are shown in Supplementary Fig. 5 and provided as a Supplemental spreadsheet, respectively. This analysis identified 3 patients with unique immune compositions and poor outcomes (supplemental information). To further explore the major drivers of variability in the data, we performed a principal component (PC) analysis. The first 2 PCs (PC-1 and PC-2) explained 15.2% and 12.5% of the variability in the data, respectively. The top and bottom 10% of immune subsets with the highest and lowest loadings for each PC are shown in Fig. 4A. Conceptually, an immune subset with high loadings within a given PC, correlates the most with that PC and is most influential in defining its immune subset composition. Consequently, patients that have high scores for this PC are expected to have higher abundance of these immune subsets. Immune subsets with high loadings within PC-1 consisted exclusively of CD4+ cells with early memory phenotypes (CD27/CD28/CD127+), whereas those with low loadings for PC-1 consisted mostly of the rare myeloid and T cell subsets that were identified during the correlation analyses described above and overlapped with most subsets with high loadings within PC-2. Finally, immune subsets with the lowest loadings for PC-2 consisted exclusively of myeloid subsets, some of which with phenotypes consistent with plasmacytoid dendritic cells (pDCs; M-26:CD123/CD303 + ) or progenitor cells (M-22, M-11: CD13/CD90/CD7/CD15 + ). These data suggest that the major drivers of variability in PBSC include subsets associated with an improved or impaired immunosurveillance (e.g., early memory CD4 subsets), various myeloid subsets, and as expected, mobilized progenitor cells.

A Immune subsets with the highest/lowest loadings for each principal component (PC). B Patients with high PC-2 scores had worse tMN-free survival, C progression-free survival from transplant and D inferior overall survival from ASCT.
Next, we analyzed these components to explore the relationships between immune system composition with clinical outcomes of interest. A high PC-2 score was associated with an inferior OS and PFS from ASCT and MNFS (Fig. 4B–D). Worse OS was also noted from MM diagnosis (not shown). Interestingly, despite the difference in outcomes, the baseline characteristics were largely comparable between the two cohorts, except for age and R-ISS stage 3 status. Patients with high PC-2 scores were older (63 vs. 59 years, P = 0.009) and were noted to have higher proportion of R-ISS stage 3 disease (54% vs. 14%, P = 0.005). In a multivariate analysis that include PC-2 score status (high vs. low), R-ISS stage 3 disease and age, PC-2 score status was not independently associated with PFS and OS from ASCT (not shown). In contrast, the high PC-2 cohort was associated with an inferior MNFS (Fig. 4B). The high PC-2 score remained an independent predictor of shorter MNFS on a multivariate analysis [HR 2.2 (95%CI: 1.1–4.4), P = 0.036] including lines of therapy received and whether lenalidomide maintenance was used or not. To understand the relationship between MNFS and MM PFS, we evaluated the extent of overlap between the development of tMN and progression of MM. Sixteen patients did not have MM progression during follow-up, of these 7 developed tMN. Thirty-eight patients had MM progression during follow-up, of which 13 also developed tMN. In these 13 cases, 12 patients developed tMN much later than the first MM progression with the exception of one patient who was diagnosed with tMN at the same time as their first MM relapse.
Immune subsets that were significantly different (FDR corrected P-value < 0.05) between the patients with high and low PC-2 scores are shown in Fig. 5. These, not unexpectedly, largely overlap with the dominant (high/low loadings) immune subsets within PC-2. Immune subsets enriched in the high PC-2 cohort included the “NKT-like” (CD56 + ) T-2 subset which expressed the inhibitory CD159a receptor [37] and TIGIT; the terminally differentiated (CD27/CD28-, CD57/KLRG1 + ) and exhausted (TIGIT/PD-1 + ) T-3 subset the exhausted (TIGIT/PD-1 + ) T-7 and T-14 subset, the immunosenescent (CD27/CD28-, CD57/KLRG1 + ) T8 subset as well as the terminally exhausted (C27/C28-, KLRG1/PD-1/TIGIT + ) TCRgd subset. Among subsets that were low within the patients with high scores for PC-2 included the HLA-DR expressing M2 subset; the CX3CR1 + M-3 subset that expressed markers of nonclassical monocytes (CD14dim/CD16 + ) and M1 polarization (CD86/CD16 +); the M-9, M-10 and M-26 subsets that expressed dendritic cell markers (CD1c/HLA-DR+ and CD123/CD303/HLA-DR + , respectively), the M-11 and M-22 subsets with a phenotype consistent with progenitor cells (CD90/CD7/CD13 + ). Given the known detrimental effects of malignant plasma cells on BM hematopoiesis [38, 39], and T cell immunosenescense [14] which could be reflected in the abundance of these subsets in PBSC, we evaluated differences in the quality of hematologic response after induction chemotherapy (VGPR or better versus not and degree of clearance of bone marrow plasma cells) between PC-2 high and PC-low patients but found no differences. These data suggest that T cell subsets associated with decreased tumor immunosurveillance are increased, whereas those associated with antigen presentation (DCs, HLA-DR + ) improved tumor outcomes (M1 monocytes) and progenitor cells are decreased in the PBSC products of patients with poor outcomes after ASCT.
<div class="c-article-section__figure js-c-reading-companion-figures-item" data-test="figure" data-container-section="figure" id="figure-5" data-title="Immune subsets significantly different (FDR p value

All p-values noted are FDR corrected.
Discussion
The utility of ASCT, a universally utilized modality in MM, is hampered by two long-term complications: relapsed MM and the development of t-MN. ASCT is often associated with high morbidity, financial toxicity, and high resource utilization. Therefore, the question of the continued relevance and timing is often raised [40]. Improved identification of patients who are likely to derive lesser benefit and/or develop t-MN may improve ASCT outcomes, while minimizing the toxicities. Given the impact of PBSCs in post-ASCT immune reconstitution, we hypothesized that the immune composition of the PBSCs can identify patients with worse outcomes.
In this study, we provide, for the first time, a comprehensive immune atlas of the lymphoid and myeloid compartment of PBSC of patients undergoing upfront ASCT for MM. We find that PBSC products enriched in immunosuppressive T and myeloid cell subsets and with a lower proportion of antigen-presenting cells, progenitor populations, and anti-tumor macrophages are associated with shorter PFS and MNFS. Specifically, the enrichment of senescent and exhausted T-cells was associated with inferior PFS/MNFS. T-cell senescence and exhaustion are known to be associated with inferior outcomes in MM at various disease stages, including at diagnosis and following ASCT [11, 17, 41]. Various factors can impact T-cell exhaustion, including chronic tumor-associated antigenic stimulation, secretion of immune suppressive cytokines by the iTME and as an effect of treatment for the malignancy. MM patients exposed to more lines of therapy develop a progressively dysfunctional iTME with relapsed/refractory MM having an abundance of senescent T-cells and decreased early memory T-cells [42]. These findings carry potential significance beyond ASCT, as these immune effector cell subsets are associated with inferior responses with subsequent T-cell redirecting therapies including bispecific antibodies and CAR-T cells [43]. Our findings suggest that these dysfunctional T-cells may arise early in the disease course and predict future inferior outcomes.
We also show that the patients with a higher proportion of exhausted T-cells had a shorter MNFS. Patients with de novo AML have high expression of inhibitory molecules (PD-1, TIGIT) on CD8 + T-cells, which in turn are associated with therapeutic resistance and disease progression [44, 45]. This immune exhausted phenotype is even more prominent in TP53mut myeloid neoplasms, which can upregulate PD-L1 expression in hematopoietic stem cells and increase the proportion of highly immunosuppressive regulatory T-Cells [19].
We observed enrichment of several differences NK-cell subsets in patients who subsequently developed a t-MN. Specifically, subsets of CD56+ expressing NK- and T-cells with the co-expression of the inhibitory receptor CD159a (NKG2A) were enriched in patients with inferior outcomes. Impaired NK-cell distribution with a phenotypic shift from a mature to immature state along with an impaired NK antitumor response in relation to the inhibitory CD159a expression has been previously demonstrated in patients with MDS and AML. The CD159a is an inhibitory immune checkpoint expressed on cytotoxic T-cells and NK-cells and upregulated in immune microenvironment of tumors treated with PD-L1 inhibitors, suggesting this to be a possible escape mechanism [46, 47]. Blockade of CD159a with targeted monoclonal antibodies has been shown to augment anti-tumor immunity as well as response to cancer vaccines and blocking CD159a may be a strategy worth exploring in patients with MM as well [48, 49]. Additionally, NK-cells with a low CD159a expression may have a role in aging-associated immune surveillance and CD159a has been shown to be upregulated in patients with ineffective erythropoiesis and MDS [37]. This may further explain the higher risk of tMN in patients enriched with CD159a expressing NKT-cells in our cohort.
Apart from being enriched in dysfunctional T-cell subsets, we show that patients with inferior MM-related outcomes and shorter tMN-free survival also had lower proportion of myeloid cells with antigen-presenting properties (HLA-DR + , pDC phenotype). Studies in precursor MM states (including MGUS and smoldering MM) have demonstrated impaired HLA-DR expression on CD14+ monocytes, which was noted to promote in vitro myeloma cell growth and suppress T-cell activation [50]. This may explain the worse MM-related survival noted in patients with lower proportion of HLA-DR expressing myeloid subsets [50]. We were able to validate the abnormal expression of CD7 as well as HLA-DR to be associated with increased t-MN risk. In patients undergoing ASCT, the presence of CD7 was associated with 6.6-fold higher risk t-MN risk [51]. Similarly, abnormal expression of CD7 and HLA-DR/CD13 was associated with 3.5- and 2.9-fold higher risk of a subsequent MN in patients with clonal cytopenia of undetermined significance (CCUS) [52].
Our findings carry implications for future therapies including CAR-T and bispecific T-cell engagers. There is ample evidence of the limited durability of the efficacy of CAR-T therapies due to the development of T-cell exhaustion and senescence [53, 54]. Pre-existing IEC dysfunction, therefore, may provide a window into assessing appropriate candidates for CAR-T therapy and further impress upon the need for developing ‘exhaustion resistant’ CARs [55, 56]. Finally, the development of t-MN after CAR-T therapy is an area of increasing concern and it is important to be able to identify patients that might have a predisposition to t-MN while assessing for CAR-T therapy. Limited data for post CAR-T t-MN suggests a somewhat overlapping transcriptional profile for t-MN developing after CAR-T therapy with de novo AML/MDS [57]. Finally, our results suggest that utilizing PBSC products as a source for CAR-T may not bypass the problem of immune dysfunction inherent in this patient population. Other sources (e.g., allogeneic CAR T) may address this issue better.
ASCT and post-ASCT lenalidomide exposure are associated with approximately 100- and 6-fold increased risk of t-MN [4, 58]. Our study suggests that the difference in immunome predates ASCT and post-ASCT lenalidomide exposure and provides a novel insight into t-MN pathogenesis. Previously, the pathogenesis of t-MN was considered a unidimensional phenomenon characterized by therapy-induced DNA damage that led to leukemic transformation of a HSC. Recently, a more nuanced model has emerged: a preleukemic clone may exist before the exposure to DNA-damaging therapies [59] and persist thereafter without manifesting as t-MN [60]. In addition, the role of non-HSC factors such as mesenchymal stromal cells has been described [61].
Factors that result in ‘tolerance’ of the clone versus progression into an overt leukemia are yet not known. Our observations suggests that the risk of t-MN predates the ASCT and maintenance therapy and point towards the importance of immune surveillance in predisposing to the development of t-MN. Conversely, the pre-existence of such anomalies paves the way for early identification, prevention, and sequencing of the available therapeutic strategies. For example, patients with such PBSC immunome could be considered for a deferred ASCT, shorter exposure to or avoidance of lenalidomide.
Limitations of our study include the selection of the study population based on the available samples rather than a pre-defined criterion. Disproportionate loss of some myeloid immune subsets remains a possibility when using cryopreserved cells. The small sample size limits the generalizability of these data which need additional validation. The immune subsets were not independently prognostic when adjusted for age and R-ISS stage. Additionally, since these phenotypes were also associated with higher R-ISS stage they may reflect a negative impact of a more aggressive tumor itself on the IEC pool [14, 62]. Finally, it is unclear if the changes noted in PBSC are also present in peripheral blood samples that would be easier to obtain. We chose to investigate PBSC since it represents the source of bone marrow engraftment after myeloablative conditioning.
Despite these limitations, our findings of the PBSC immunome composition predicting inferior MM-related outcomes and future risk of tMN will guide future confirmatory studies. The use of a wide array of therapies and the protracted follow-up needed make such studies challenging. Therefore, our study is uniquely positioned to answer the question of the impact of the PBSC immunome on future risk of relapse and tMN development.
Data availability
The raw data from the mass cytometry experiments are available at www.flowrepository.org.
References
-
Binder M, Nandakumar B, Rajkumar SV, Kapoor P, Buadi FK, Dingli D, et al. Mortality trends in multiple myeloma after the introduction of novel therapies in the United States. Leukemia. 2022;36:801–8.
Google Scholar
-
Richardson PG, Jacobus SJ, Weller EA, Hassoun H, Lonial S, Raje NS, et al. Triplet therapy, transplantation, and maintenance until progression in myeloma. N Engl J Med. 2022;387:132–47.
Google Scholar
-
Sperling AS, Guerra VA, Kennedy JA, Yan Y, Hsu JI, Wang F, et al. Lenalidomide promotes the development of TP53-mutated therapy-related myeloid neoplasms. Blood. 2022;140:1753–63.
Google Scholar
-
Nadiminti K, Sidiqi MH, Meleveedu K, Alkhateeb HB, Hogan WJ, Litzow M, et al. Characteristics and outcomes of therapy-related myeloid neoplasms following autologous stem cell transplantation for multiple myeloma. Blood Cancer J. 2021;11:63.
Google Scholar
-
Ragon BK, Shah MV, D’Souza A, Estrada-Merly N, Gowda L, George G, et al. Impact of second primary malignancy post-autologous transplantation on outcomes of multiple myeloma: a CIBMTR analysis. Blood Adv. 2023;7:2746–57.
Google Scholar
-
Hiwase D, Hahn C, Tran ENH, Chhetri R, Baranwal A, Al-Kali A, et al. TP53 mutation in therapy-related myeloid neoplasm defines a distinct molecular subtype. Blood. 2023;141:1087–91.
Google Scholar
-
Thibaud S, Mia MB, Van Oekelen O, Mouhieddine TH, Schaniel C, Ghodke-Puranik Y, et al. Comprehensive characterization of prolonged unexplained cytopenias in relapsed/refractory multiple myeloma patients following BCMA-directed CAR-T cell therapy. Blood. 2022;140:614–6.
Google Scholar
-
Alkhateeb HB, Mohty R, Greipp P, Bansal R, Hathcock M, Rosenthal A, et al. Therapy-related myeloid neoplasms following chimeric antigen receptor T-cell therapy for Non-Hodgkin Lymphoma. Blood Cancer J. 2022;12:113.
Google Scholar
-
Zanwar S, Nandakumar B, Kumar S. Immune-based therapies in the management of multiple myeloma. Blood Cancer J. 2020;10:84.
Google Scholar
-
Bianchi G, Munshi NC. Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood. 2015;125:3049–58.
Google Scholar
-
Kourelis TV, Villasboas JC, Jessen E, Dasari S, Dispenzieri A, Jevremovic D, et al. Mass cytometry dissects T cell heterogeneity in the immune tumor microenvironment of common dysproteinemias at diagnosis and after first line therapies. Blood Cancer J. 2019;9:72.
Google Scholar
-
Pilcher W, Thomas BE, Bhasin SS, Jayasinghe RG, Yao L, Gonzalez-Kozlova E, et al. Cross center single-cell RNA sequencing study of the immune microenvironment in rapid progressing multiple myeloma. NPJ Genom Med. 2023;8:3.
Google Scholar
-
Arteche-López A, Kreutzman A, Alegre A, Sanz Martín P, Aguado B, González-Pardo M, et al. Multiple myeloma patients in long-term complete response after autologous stem cell transplantation express a particular immune signature with potential prognostic implication. Bone Marrow Transpl. 2017;52:832–8.
Google Scholar
-
Suen H, Brown R, Yang S, Weatherburn C, Ho PJ, Woodland N, et al. Multiple myeloma causes clonal T-cell immunosenescence: identification of potential novel targets for promoting tumour immunity and implications for checkpoint blockade. Leukemia. 2016;30:1716–24.
Google Scholar
-
Chung DJ, Pronschinske KB, Shyer JA, Sharma S, Leung S, Curran SA, et al. T-cell exhaustion in multiple myeloma relapse after autotransplant: optimal timing of immunotherapy. Cancer Immunol Res. 2016;4:61–71.
Google Scholar
-
Friedrich MJ, Neri P, Kehl N, Michel J, Steiger S, Kilian M, et al. The pre-existing T cell landscape determines the response to bispecific T cell engagers in multiple myeloma patients. Cancer Cell. 2023;41:711–25.e716.
Google Scholar
-
Parmar H, Gertz M, Anderson EI, Kumar S, Kourelis TV. Microenvironment immune reconstitution patterns correlate with outcomes after autologous transplant in multiple myeloma. Blood Adv. 2021;5:1797–804.
Google Scholar
-
Seggewiss R, Einsele H. Immune reconstitution after allogeneic transplantation and expanding options for immunomodulation: an update. Blood. 2010;115:3861–8.
Google Scholar
-
Sallman DA, McLemore AF, Aldrich AL, Komrokji RS, McGraw KL, Dhawan A, et al. TP53 mutations in myelodysplastic syndromes and secondary AML confer an immunosuppressive phenotype. Blood. 2020;136:2812–23.
Google Scholar
-
Yu S, Ren X, Meng F, Guo X, Tao J, Zhang W, et al. TIM3/CEACAM1 pathway involves in myeloid-derived suppressor cells induced CD8(+) T cells exhaustion and bone marrow inflammatory microenvironment in myelodysplastic syndrome. Immunology. 2023;168:273–89.
Google Scholar
-
Swatler J, Turos-Korgul L, Kozlowska E, Piwocka K. Immunosuppressive cell subsets and factors in myeloid leukemias. Cancers (Basel). 2021;13:1203.
Google Scholar
-
Kong Y, Zhu L, Schell TD, Zhang J, Claxton DF, Ehmann WC, et al. T-Cell immunoglobulin and ITIM domain (TIGIT) associates with CD8+ T-cell exhaustion and poor clinical outcome in AML patients. Clin Cancer Res. 2016;22:3057–66.
Google Scholar
-
Epling-Burnette PK, Painter JS, Rollison DE, Ku E, Vendron D, Widen R, et al. Prevalence and clinical association of clonal T-cell expansions in Myelodysplastic Syndrome. Leukemia. 2007;21:659–67.
Google Scholar
-
Kook H, Zeng W, Guibin C, Kirby M, Young NS, Maciejewski JP. Increased cytotoxic T cells with effector phenotype in aplastic anemia and myelodysplasia. Exp Hematol. 2001;29:1270–7.
Google Scholar
-
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405.
Google Scholar
-
Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J Clin Oncol. 2015;33:2863–9.
Google Scholar
-
Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17:e328–e346.
Google Scholar
-
Maiolino A, Biasoli I, Lima J, Portugal AC, Pulcheri W, Nucci M. Engraftment syndrome following autologous hematopoietic stem cell transplantation: definition of diagnostic criteria. Bone Marrow Transplant. 2003;31:393–7. 2003/03/01
Google Scholar
-
Hahne F, Khodabakhshi AH, Bashashati A, Wong CJ, Gascoyne RD, Weng AP, et al. Per-channel basis normalization methods for flow cytometry data. Cytom A. 2010;77:121–31.
-
Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(Jul):184–97.
Google Scholar
-
Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44.
Google Scholar
-
Cianga VA, Campos Catafal L, Cianga P, Pavel Tanasa M, Cherry M, Collet P, et al. Natural killer cell subpopulations and inhibitory receptor dynamics in myelodysplastic syndromes and acute myeloid leukemia. Front Immunol. 2021;12:665541.
Google Scholar
-
Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. 2019;10:3931.
Google Scholar
-
Hamann I, Unterwalder N, Cardona AE, Meisel C, Zipp F, Ransohoff RM, et al. Analyses of phenotypic and functional characteristics of CX3CR1-expressing natural killer cells. Immunology. 2011;133:62–73.
Google Scholar
-
Ali A, Canaday LM, Feldman HA, Cevik H, Moran MT, Rajaram S, et al. Natural killer cell immunosuppressive function requires CXCR3-dependent redistribution within lymphoid tissues. J Clin Invest. 2021;131. https://doi.org/10.1172/JCI146686.
-
Goedhart M, Gessel S, van der Voort R, Slot E, Lucas B, Gielen E, et al. CXCR4, but not CXCR3, drives CD8(+) T-cell entry into and migration through the murine bone marrow. Eur J Immunol. 2019;49:576–89.
Google Scholar
-
Manser AR, Uhrberg M. Age-related changes in natural killer cell repertoires: impact on NK cell function and immune surveillance. Cancer Immunol Immunother. 2016;65:417–26.
Google Scholar
-
Bruns I, Cadeddu RP, Brueckmann I, Fröbel J, Geyh S, Büst S, et al. Multiple myeloma-related deregulation of bone marrow-derived CD34(+) hematopoietic stem and progenitor cells. Blood. 2012;120:2620–30.
Google Scholar
-
Laurenzana I, Trino S, Lamorte D, De Stradis A, Santodirocco M, Sgambato A, et al. Multiple myeloma-derived extracellular vesicles impair normal hematopoiesis by acting on hematopoietic stem and progenitor cells. Front Med (Lausanne). 2021;8:793040.
Google Scholar
-
Kumar SK, Buadi FK, Rajkumar SV. Pros and cons of frontline autologous transplant in multiple myeloma: the debate over timing. Blood. 2019;133:652–9.
Google Scholar
-
Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792.
Google Scholar
-
Visram A, Dasari S, Anderson E, Kumar S, Kourelis TV. Relapsed multiple myeloma demonstrates distinct patterns of immune microenvironment and malignant cell-mediated immunosuppression. Blood Cancer J. 2021;11:45.
Google Scholar
-
Majzner RG, Mackall CL. Clinical lessons learned from the first leg of the CAR T cell journey. Nat Med. 2019;25:1341–55.
Google Scholar
-
Zhou Q, Munger ME, Veenstra RG, Weigel BJ, Hirashima M, Munn DH, et al. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117:4501–10.
Google Scholar
-
Wang M, Bu J, Zhou M, Sido J, Lin Y, Liu G, et al. CD8(+)T cells expressing both PD-1 and TIGIT but not CD226 are dysfunctional in acute myeloid leukemia (AML) patients. Clin Immunol. 2018;190:64–73.
Google Scholar
-
Creelan BC, Antonia SJ. The NKG2A immune checkpoint – a new direction in cancer immunotherapy. Nat Rev Clin Oncol. 2019;16:277–8.
Google Scholar
-
Mingari MC, Pietra G, Moretta L. Immune checkpoint inhibitors: anti-NKG2A antibodies on board. Trends Immunol. 2019;40:83–85.
Google Scholar
-
van Montfoort N, Borst L, Korrer MJ, Sluijter M, Marijt KA, Santegoets SJ, et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell. 2018;175:1744–55.
Google Scholar
-
André P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175:1731–43.
Google Scholar
-
Zavidij O, Haradhvala NJ, Mouhieddine TH, Sklavenitis-Pistofidis R, Cai S, Reidy M, et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat Cancer. 2020;1:493–506.
Google Scholar
-
Soerensen JF, Aggerholm A, Kerndrup GB, Hansen MC, Ewald IKL, Bill M, et al. Clonal hematopoiesis predicts development of therapy-related myeloid neoplasms post-autologous stem cell transplantation. Blood Adv. 2020;4:885–92.
Google Scholar
-
Jevremovic D, Nanaa A, Geyer SM, Timm M, Azouz H, Hengel C, et al. Abnormal CD13/HLA-DR expression pattern on myeloblasts predicts development of myeloid neoplasia in patients with clonal cytopenia of undetermined significance. Am J Clin Pathol. 2022;158:530–6.
Google Scholar
-
Poorebrahim M, Melief J, Pico de Coaña Y L, Wickström S, Cid-Arregui A, Kiessling R. Counteracting CAR T cell dysfunction. Oncogene. 2021;40:421–35. 2021/01/01
Google Scholar
-
Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol. 2019;16:372–85.
Google Scholar
-
Maus MV. Immunology: T-cell tweaks to target tumours. Nature. 2017;543:48–49.
Google Scholar
-
Jung IY, Lee J. Unleashing the therapeutic potential of CAR-T cell therapy using gene-editing technologies. Mol Cells. 2018;41:717–23.
Google Scholar
-
Zhao A, Zhao M, Qian W, Liang A, Li P, Liu H. Secondary myeloid neoplasms after CD19 CAR T therapy in patients with refractory/relapsed B-cell lymphoma: case series and review of literature. Front Immunol. 2022;13:1063986.
Google Scholar
-
Radivoyevitch T, Dean RM, Shaw BE, Brazauskas R, Tecca HR, Molenaar RJ, et al. Risk of acute myeloid leukemia and myelodysplastic syndrome after autotransplants for lymphomas and plasma cell myeloma. Leuk Res. 2018;74:130–6.
Google Scholar
-
Young AL, Wong TN, Hughes AEO, Heath SE, Ley TJ, Link DC, et al. Quantifying ultra-rare pre-leukemic clones via targeted error-corrected sequencing. Leukemia. 2015;29:1608–11.
Google Scholar
-
Shah MV, Mangaonkar AA, Begna KH, Alkhateeb HB, Greipp P, Nanaa A, et al. Therapy-related clonal cytopenia as a precursor to therapy-related myeloid neoplasms. Blood Cancer J. 2022;12:106.
Google Scholar
-
Kutyna MM, Kok CH, Lim Y, Tran ENH, Campbell D, Paton S, et al. A senescence stress secretome is a hallmark of therapy-related myeloid neoplasm stromal tissue occurring soon after cytotoxic exposure. Leukemia. 2022;36:2678–89.
Google Scholar
-
Zhang Y, Pfannenstiel LW, Bolesta E, Montes CL, Zhang X, Chapoval AI, et al. Interleukin-7 inhibits tumor-induced CD27-CD28- suppressor T cells: implications for cancer immunotherapy. Clin Cancer Res. 2011;17:4975–86.
Google Scholar
Author information
Authors and Affiliations
Contributions
SZ, MVS, TK conceived the study, collected the data, performed the preliminary analysis and wrote the initial draft of the manuscript. EJ, CG processed the stem cell product and critically appraised the manuscript. KP and MS performed the staining and processing of samples for mass cytometry. EA, AA, MW critically appraised the manuscript and suggested modifications. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
MVS reports institutional research funding from Bristol-Meyers Squibb, AbbVie, Astellas, and MRKR Therapeutics. The remaining authors do not have any relevant conflicts of interest to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Correlation Matrix Supplementary Sheet
Supplementary Material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Zanwar, S., Jacob, E.K., Greiner, C. et al. The immunome of mobilized peripheral blood stem cells is predictive of long-term outcomes and therapy-related myeloid neoplasms in patients with multiple myeloma undergoing autologous stem cell transplant.
Blood Cancer J. 13, 151 (2023). https://doi.org/10.1038/s41408-023-00920-9
-
Received: 12 June 2023
-
Revised: 22 August 2023
-
Accepted: 01 September 2023
-
Published: 26 September 2023
-
DOI: https://doi.org/10.1038/s41408-023-00920-9