Cancer and neoplasms
MALAT1 expression is associated with aggressive behavior in indolent B-cell neoplasms
Abstract
MALAT1 long non-coding RNA has oncogenic roles but has been poorly studied in indolent B-cell neoplasms. Here, MALAT1 expression was analyzed using RNA-seq, microarrays or qRT-PCR in primary samples from clinico-biological subtypes of chronic lymphocytic leukemia (CLL, n = 266), paired Richter transformation (RT, n = 6) and follicular lymphoma (FL, n = 61). In peripheral blood (PB) CLL samples, high MALAT1 expression was associated with a significantly shorter time to treatment independently from other known prognostic factors. Coding genes expressed in association with MALAT1 in CLL were predominantly related to oncogenic pathways stimulated in the lymph node (LN) microenvironment. In RT paired samples, MALAT1 levels were lower, concordant with their acquired increased independency of external signals. Moreover, MALAT1 levels in paired PB/LN CLLs were similar, suggesting that the prognostic value of MALAT1 expression in PB is mirroring expression differences already present in LN. Similarly, high MALAT1 expression in FL predicted for a shorter progression-free survival, in association with expression pathways promoting FL pathogenesis. In summary, MALAT1 expression is related to pathophysiology and more aggressive clinical behavior of indolent B-cell neoplasms. Particularly in CLL, its levels could be a surrogate marker of the microenvironment stimulation and may contribute to refine the clinical management of these patients.
Introduction
Long non-coding RNAs (lncRNAs) regulate the expression of protein-coding genes and are increasingly described as key players in physiological and pathological conditions, most remarkably in cancer1,2. One of the lncRNAs most frequently related to oncogenesis is MALAT1, which has been implicated in the regulation of key cellular pathways such as MAPK/ERK, PI3K/AKT, WNT/B-catenin, and NF-kB3 and, as a consequence, involved in many cancer-associated processes such as cell proliferation, migration, invasion, apoptosis and angiogenesis4. There are also several studies supporting MALAT1 expression as a clinical biomarker mainly associated with a poor prognosis in solid tumors3, although in some neoplasms such as diffuse large B-cell lymphoma (DLBCL), colorectal and breast cancer, high levels of MALAT1 have been linked to a favorable outcome5,6,7.
In B-cell non-Hodgkin lymphomas (B-NHL), we and others have previously shown that lncRNA deregulation is associated with immune cell-related functions and cell proliferation control8,9. In the case of MALAT1, its expression has been shown to be upregulated in some lymphoid neoplasms such as DLBCL10, chronic lymphocytic leukemia (CLL)11, and mantle cell lymphoma (MCL)12. These results suggest that MALAT1 expression could be associated with poor prognosis even in indolent B-NHL, but most of these studies include relatively few cases and the possible clinical relevance of MALAT1 expression in these lymphoid neoplasms is still not well known. In this study, we have performed a detailed characterization of the biological and clinical impact of MALAT1 deregulation in two types of indolent B cell lymphomas with different underlying pathobiological mechanisms, namely CLL and follicular lymphoma (FL).
Results
MALAT1 expression in CLL
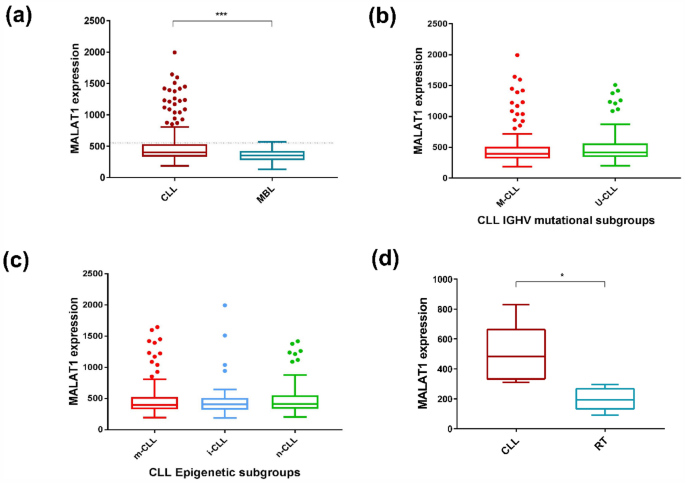
We initially evaluated the expression levels of MALAT1 in 266 CLL (Cohort CLL#1) and 25 monoclonal B cell lymphocytosis (MBL) (Table 1). MALAT1 expression was significantly higher in CLL than in MBL (P < 0.001) (Fig. 1a). We also stratified MALAT1 levels according to the IGHV mutational status (mutated -M-CLL- vs un-mutated U-CLL) and epigenetic subtypes (naïve-like -n-CLL, memory-like -m-CLL and intermediate -i-CLL-) and no significant differences were observed among the different groups (Fig. 1b and c). In addition, we analyzed MALAT1 expression in 6 paired CLL and their clonally related transformation to diffuse large B cell lymphoma (Richter transformation, RT) (CLL#2 series, Table 1). MALAT1 was significantly downregulated in the RT samples (P < 0.01) (Fig. 1d).
MALAT1 expression levels in MBL, CLL#1 molecular subgroups, and RT. (a) MALAT1 expression was higher in CLL patients than in MBL (P < 0.001). Dotted line indicates the threshold of MALAT1 that determines differences in outcome. (b) No significant differences were observed in MALAT1 expression between CLL subgroups defined by IGHV mutational status (M-CLL versus U-CLL). (c) No significant differences were observed in MALAT1 expression between CLL subgroups defined by epigenetic subtypes (n-CLL: naïve-like, m-CLL: memory-like and i-CLL: intermediate) (right panel). (d) MALAT1 expression was higher in a series of 6 samples of CLL patients than in their corresponding paired RT samples (P < 0.01).
We next evaluated whether MALAT1 expression was associated with the clinical behavior of the tumor. Using the maxstat algorithm, we segregated CLL patients into high and low expression groups (Supplementary Fig. S1a online). Patients with high MALAT1 expression (N = 65) had a significantly shorter time to treatment (TTT) than the MALAT1-low group (N = 201) (p < 0.0001) (Fig. 2a). This finding was also confirmed in the subset of patients clinically classified as Binet A (N = 239, 57 with high and 182 low MALAT1 levels) (P = 0.0003) (Supplementary Fig. S2a online). On the contrary, MALAT1 levels were not related to the overall survival (OS) of the patients in the whole series (Supplementary Fig. S1b and S2b online) or stratifying the patients according to the use (N = 77) or not (N = 60) of rituximab on different chemotherapy regimens (Supplementary Table S1 online) (P = 0.618 and P = 0.758, respectively, data not shown). The adverse impact of high MALAT1 expression on TTT was further confirmed in IGHV-mutated CLL and in the three epigenetic CLL subtypes (Fig. 2a). Similar findings were observed when the analyses were restricted to patients with Binet A CLL (Supplementary Fig. S2c,d online). To evaluate the independent prognostic impact of MALAT1, we performed multivariate regression models. First, we checked that MALAT1 as a continuous isolated variable also had significant prognostic value for TTT (HR = 1.32; 95%CI: 1.18–1.48; p < 0.0001) but not for OS (HR = 1.05; 95%CI: 0.89–1.24; p = 0.561). Next, the multivariate analyses confirmed that MALAT1 expression also had an independent prognostic value for TTT considering Binet stage and the IGHV mutational status (P = 0.0004) and Binet stage and the epigenetic subgroups (P < 0.0001) (Fig. 2b). We also evaluated the possible association of MALAT1 levels with other molecular factors previously shown to have prognostic value in CLL, such as mutations in driver genes, number of chromosomal aberrations13,14, IGLV3-21 variant/R110 mutation15, or the DNA methylation-based epiCMIT score related to the proliferative history of the tumor cells16. MALAT1 expression levels were not related to any of these variables (Supplementary Fig. S3 and S4 online). Overall, these data indicate that MALAT1 expression has a prognostic value for TTT in CLL irrespective of other known genetic and epigenetic prognostic parameters.

MALAT1 expression is an independent prognostic factor for time to treatment (TTT) in CLL. (a) CLL patients with high MALAT1 expression showed significantly shorter TTT compared to those with low levels in the global cohort (top panel) and almost every one of the different CLL subgroups related to the IGHV mutational status (bottom left) and epigenetic subtypes (bottom right). (b) Plot for multivariate model analyses, where MALAT1 expression as a continuous variable shows its independent prognostic value considered together with Binet stage and IGHV mutational status (left panel) or epigenetic subtypes (right panel). For epigenetic groups, m-CLL was taken as a reference.
To determine whether MALAT1 deregulation could be related to genetic alterations, we analyzed the gene mutational status in the whole-genome sequences of 150 cases from the ICGC Consortium13. Only five CLL and one MBL revealed mutations in the MALAT1 locus and were not related to the expression of the gene (Supplementary Table S2a online). No copy number alterations affecting 11q13, where MALAT1 is located, were found, whereas only 2 CLL cases had copy number neutral loss of heterozygosity. None of these alterations were related to the expression of the gene (Supplementary Table S2b online). We also consider the possible association of MALAT1 expression with the methylation status of this gene using previously published data included in the CLL#1 series16. The CpGs with methylation status available in the MALAT1 gene region were mainly hypomethylated, but using clustering analysis we could not observe any association between CpG status and MALAT1 expression, neither considered as a continuous nor categorical variable in any of the different CLL subtypes (Supplementary Fig. S5 online).
To evaluate the possible functional implications of MALAT1 expression we searched for coding genes that were significantly correlated, either positively or negatively, with MALAT1 in the different CLL subgroups (M-CLL and U-CLL and epigenetic subtypes) (Supplementary Table S3 online) and we subsequently performed pathway enrichment analyses. The positively correlated genes were significantly enriched in a signature highly expressed in nodal CLL compared to their respective peripheral blood sample. This signature was enriched in both U-CLL and M-CLL (Fig. 3a)17. Other pathways identified were related to activation, proliferation, and survival of CLL cells in the lymph node (LN) microenvironment, particularly PI3K/AKT, MAPK, and signaling of several interleukins, among others18,19,20,21,22 (Fig. 3b, Supplementary Fig. S6a and Table S4 online). The top core genes enriched in these pathways had been previously related to progression of the cell cycle as NDEL1, PAFAH1B1 and PDS5A23,24,25,26, or involved in cytokine-related pathways such as CCL3 and PTGS2 (Fig. 3b and Supplementary Table S5 online). CCL3 has been critically related to the composition of LN microenvironment promoting CLL survival and proliferation27. PTGS2 is already known to be overexpressed in CLL promoting resistance to apoptosis28 and being induced by different factors through STAT3 activation29,30. Other noticeable examples were PIK3CB, related to PI3K/AKT and MAPK pathways31, RHEB, a GTPase upstream activator of MTOR that cooperates with PTEN haploinsufficiency in MYC-induced murine lymphomas32, and GLS in the TP53 transcriptional regulation pathway, overexpressed in many cancers33. Moreover, in the MAPK pathway we observed a high correlation with MAPK6 whose overexpression has been associated with poor survival in several solid tumors34.

MALAT1 levels correlate positively with genes enriched in survival and proliferation pathways, including a signature described as upregulated in LN compared to PB CLL samples. (a) GSEA analysis shows that a nodal CLL signature (see material and methods) was significantly enriched in coding genes correlated with MALAT1 expression in PB CLL samples of M-CLL and U-CLL subtypes. (b) Summary of most relevant significant pathway enrichments found using Metascape tool (left panels) regarding coding genes positively (top panels) and negatively (bottom panels) correlated with MALAT1 expression in CLL subtypes defined by IGHV mutational status. Only statistically significant Reactome pathways after multiple comparison correction are shown for compact representation. Correlation values of the core enriched genes of the corresponding pathways are depicted in the right panels with relevant genes marked in colored asterisks according to the degree of correlation (in blue those more highly correlated with MALAT1).
On the other hand, the analysis of the pathways and their top genes negatively correlated with MALAT1 showed significantly enrichment of DNA damage and repair pathway (e.g. GPS1, a gene described as a suppressor of mitogen-activated signal transduction in addition to the impairing in DNA repair induced by its depletion)35, TNFR signaling (SHARPIN, a negative regulator of NF-kB via TNFR) and TP53 mediated transcriptional regulation (PIN1, an inhibitor of TP53 activation) (Fig. 3b, Supplementary Table S5 and S6 online)36,37.
We subsequently analyzed the differentially expressed genes between cases with high and low MALAT1 expression in U-CLL and M-CLL of the CLL#1 series. A total of 35 (M-CLL) and 65 genes (U-CLL) were found with significant differences in expression (Supplementary Fig. S7 and Table S7 online). These genes include several identified in the previous pathway analysis of correlated genes with MALAT1 expression and were related to the influence of the microenvironment in the lymph node (e.g. FOSB, DUSP1, CD69, NR4A2 and KLF10) (Supplementary Table S8 online)17. To further understand the significance of the differentially expressed genes we performed a GSEA analyses on genes ranked by the degree of differential expression. This analysis confirmed pathways identified in the previous analysis of genes correlated with MALAT1 expression such as the signature highly expressed in nodal CLL and cell cycle regulation, as well as pathways related to regulation of proliferation and NF-kB activation, among others involved in CLL pathogenesis (Fig. 4a and Supplementary Table S9). We also detected a negative enrichment of OXPHOS-related pathways, particularly for U-CLL (Fig. 4b and Supplementary Table S9 online). Interestingly, this later observation was concordant with the significant MALAT1 downregulation that we observed in the RT samples, since an increased OXPHOS pathway has been recently identified as characteristic of RT38.

Pathways enriched by GSEA among the genes ranked by their degree of differential expression between high and low MALAT1 expression groups in CLL IGHV subtypes. (a) Positive significantly enriched pathways related to survival, proliferation and a signature of genes described as upregulated in LN vs PB CLL samples. (b) Negative significantly enriched pathways related to OXPHOS.
Finally, we also analyzed previously published microarray data of paired LN or bone marrow (BM) and PB CLL samples39 (CLL#3 series, Table 1), where we found that MALAT1 levels did not change significantly between LN or BM compared to PB (P = 0.097; P = 0.698, respectively) (Supplementary Fig. S8 online). All these results suggest that the relationship between MALAT1 expression and shorter TTT that we have observed in PB samples could represent a surrogate biomarker for the degree of stimulation and proliferation of CLL cells in the LN.
MALAT1 expression in FL
Based on the results of MALAT1 in CLL and its potential association with the LN microenvironment, we further explored the role of MALAT1 in FL, another indolent B cell lymphoid neoplasm with predominant nodal presentation. MALAT1 expression levels were analyzed by qRT-PCR in a series of 61 grade 1-3A FL (FL#1 series, Table 1). These patients had been homogenously treated with R-CHOP. Using the maxstat algorithm, we identified two groups of FL with high (N = 11) and low (N = 50) MALAT1 expression. FL cases with high MALAT1 expression had a significantly shorter progression-free survival (PFS) than cases with low expression (P = 0.017) (Fig. 5a). However, no significant differences were observed in transformation to DLBCL or OS between cases with low and high expression (P = 0.088 and P = 0.0177) (Supplementary Fig. S9 online). Other clinical, biological, and histological characteristics were similar in FL with high and low MALAT1 expression, including parameters of known prognostic impact in FL such as high-risk FLIPI, stage or histological grade (Supplementary Table S10 online), suggesting that the prognostic value of MALAT1 is independent of these parameters.

MALAT1 expression is associated with a more aggressive behavior and is related to pathogenetic pathways in FL. (a) Kaplan–Meier curves for progression-free survival (PFS) according to MALAT1 expression in FL. Patients with high MALAT1 expression had a significantly shorter PFS than those with low levels. (b) Summary of most relevant significant pathway enrichments found with Metascape tool among coding genes positively correlated with MALAT1 in FL samples (left panel). Only statistically significant Reactome pathways after multiple comparison correction are shown for compact representation. Correlation values of the core enriched genes of the corresponding pathways are depicted in the right panel with relevant genes marked in colored asterisks according to the degree of correlation (in blue those more highly correlated with MALAT1).
To explore the possible biological role of MALAT1 expression in FL we reanalyzed microarray expression data previously generated on purified FL B-cells40. We observed that the coding genes positively correlating with MALAT1 expression (Supplementary Table S11 online) were associated with cell proliferation and other pathways like VEGF, MAPK, interleukin signaling (including some particular pathways related to IL-3 and IL-5) and BCR signaling, also described as involved in FL oncogenesis (Fig. 5b and Supplementary Table S12 online)41,42. Moreover, among the core genes positively correlated with MALAT1 we noticed several genes which function is concordant with the aggressiveness of high MALAT1 levels observed in FL (Fig. 5b and Supplementary Table S13 online). In this way, PIK3CD, LYN and GRB2 were involved in several pathways and PIK3CD was previously described as predictor of poor prognosis in FL43. We also found others genes related to cytokine and interleukin signaling and other pathways involved in tumor progression (Fig. 5b)44,45,46,47,48,49.
In a subsequent analysis of differentially expressed genes between FL with high and low MALAT1 expression in FL#2 cohort we identified 26 genes (Supplementary Fig. S10 and Table S14 online). The top overexpressed gene was IL-7 which high expression has been previously related to the activation of Tfh cells in FL50. The second overexpressed gene was GNG11 previously found overexpressed in FL with shorter response to R-CHOP therapy51. The GSEA analysis on the global gene list ranked by differential expression did not show relevant pathways related to FL.
Finally, we performed a comparison of all genes involved in pathways significantly enriched in FL and compared to those found in CLL (considering together U-CLL and M-CLL gene lists). A large proportion of those genes were exclusive for each lymphoma type (88% in FL and 99% in CLL) (Supplementary Fig. S11 and Table S15 online). Concordantly, cell cycle-related or MAPK pathways that were initially observed in both neoplasms involved different sets of MALAT1 positively correlated genes (Supplementary Tables S16 and S17 online). On the other hand, a total of 12 genes were found in common in the comparative analysis, including several involved in cytokine and interleukin signaling (CAPZA1), cell cycle (GAR1), or both (HSP90AA1, LMNB1 and LYN)(Supplementary Table S18 online).
Discussion
In this report, we provide novel insights into the clinical and biological role of MALAT1 in indolent B cell neoplasms. Our study revealed that MALAT1 upregulation was associated with a detrimental clinical behavior in the different entities studied, associated with a shorter TTT in CLL or shorter PFS in FL. Interestingly, MALAT1 upregulation was a prognostic factor in CLL independently of the IGHV mutational status, epigenetic subgroups or Binet stage. In FL, MALAT1 overexpression was also associated with shorter progression-free survival although the clinical and biological features of patients with low and high expression were similar.
We also studied the potential causes and consequences of MALAT1 upregulation that might justify its clinical behavior in indolent B cell neoplasms. In CLL, MALAT1 mutations were rare and not related to the expression of the gene. This finding is similar to those in other neoplasms such as bladder cancer, head and neck squamous cell cancer, and lung adenocarcinomas, in which single nucleotides variants and indels were found, although considered passenger events caused by a transcription-associated mutational process52,53. Moreover, we also considered if changes in the methylation status of MALAT1 could be associated with its upregulation as previously showed in lung cancer54. However, the region studied in CLL was found mainly hypomethylated and without any clear association with MALAT1 expression differences across patients.
Therefore, other mechanisms should be related to MALAT1 overexpression. In this sense, the transcription factors HIF1α and STAT3 induced by microenvironmental factors such as hypoxia and cytokines/interleukins bind to MALAT1 promoter increasing its expression55,56,57,58. In CLL, microenvironment stimulation occurs in the LN and is a key process for activation, proliferation and survival of these tumoral B cells59. Interestingly, our guilt-by-association analysis on the coding genes correlated with MALAT1 in CLL as well as the differentially expressed genes in cases with high and low MALAT1 revealed significant enrichment in genes signature highly expressed in nodal CLL17, and interleukin/cytokine-signalling27, suggesting a close relationship between MALAT1 expression and the influence of the nodal microenvironment. Interestingly, we found that MALAT1 levels remained similar in PB compared to LN/BM paired CLL samples, supporting that the clinical value of MALAT1 expression found in PB is mirroring expression differences in other compartments as in the LN, and that could be associated with the degree of the microenvironment stimulation. Other pathways upregulated in relation to MALAT1 overexpression were mainly involved in cell cycle regulation and signal transduction, also known to be stimulated by nodal microenvironment and related to CLL aggressiveness22,60.
Intriguingly, we observed MALAT1 downregulation in the aggressive RT compared to their paired CLL phase. This result could be related to the increased genomic alterations acquired in RT that may drive proliferation and survival of tumor cells more independently of the microenvironment38,61. Interestingly, one of the pathways downregulated in association with high MALAT1 expression in CLL was OXPHOS. These findings are concordant with the high OXPHOS activity recently identified as characteristic of RT together with downregulation of the BCR signaling as additional evidence of increasing independence of transformed tumor cells from the microenvironment stimuli38.
MALAT1 expression in CLL was not related to any of the major genomic alteration with known prognostic impact in the evolution of the disease. This finding is concordant with the predictive value of MALAT1 overexpression for shorter TTT together with the mutational status of the IGHV and Binet stage, independently of strong driver alterations, including TP53 mutations or deletions (see Supplementary Fig. 3b,c online). However, the pathway analysis of the genes correlated to MALAT1 expression showed that one of the significant altered pathways was transcriptional regulation of TP53 (see Fig. 3b). One of the genes downregulated in association with MALAT1 was PIN1 that seems to be required for the transcriptional activity of wild-type TP5337, suggesting that the low levels of PIN1 found in CLL with high MALAT1 expression could contribute to impair TP53 activity even in absence of alterations in this tumor suppressor gene.
In FL, our study also revealed biological insights that seem to explain the clinical behavior of MALAT1 upregulation in these neoplasms and its relationship to the aggressiveness of the tumors. Coding genes correlated with MALAT1 expression in FL were enriched in cell cycle-related processes as well as pathways related to cell proliferation, migration and angiogenesis, such as MAPK and VEGF pathways, which have been previously linked to MALAT1 function in other tumor models62. We also found that high MALAT1 expression was significantly associated with high levels of genes involved in several other pathways as BCR and interleukin signaling, such as LYN and PIK3CD, the expression of which promote cell growth and have been associated with poor prognosis in FL43,63. Moreover, the study of differentially expressed genes between case with high and low MALAT1 identified IL-7 previously described as upregulated in FL B-cells paralleling the levels of activation of Tfh cells50 and GNG11 highly expressed in FL with short response to R-CHOP therapy51. All these are relevant associations supporting the clinical impact of MALAT1 in FL even though our series included a relative limited number of cases.
Finally, the comparison of MALAT1 associated pathways and the involved genes showed a low degree of overlapping in the corresponding gene signatures, even in cell cycle-related or MAPK pathways that were initially observed as enriched in both neoplasms. These results suggest that MALAT1 could be contributing to the regulation of different transcriptional gene sets in both lymphomas. In spite of these differences, we also observed that processes as cell proliferation and MAPK pathways were potentially commonly affected by MALAT1 upregulation, and thus could explain its common clinical impact in both neoplasms.
In summary, these findings highlight that MALAT1 overexpression plays a role in the pathobiology and clinical behavior of indolent B cell neoplasms related to a more aggressive behavior of the tumors with higher MALAT1 levels. Particularly in CLL, MALAT1 could serve as a clinical biomarker that seems to be a surrogate indicator of the degree of stimuli the CLL cells receive from the microenvironment. Therefore, MALAT1 expression could be taken in account in further studies as complementary to other known prognostic factors to improve the clinical management of these patients.
Material and methods
Description of the transcriptional data and patient cohorts/features
This study exploited previously published genome wide transcriptional data from CLL and FL. Regarding CLL, we reanalyzed our RNA-seq data from PB pre-treatment samples obtained from 266 patients, and 25 additional monoclonal B cell lymphocytosis donors (MBL) of the Spanish ICGC consortium with full clinical annotations13 (MBL and CLL#1 series, Table 1). The CLL subtypes according to IGHV mutational status and epigenetic subtypes were identified following standard methods published elsewhere64,65. MALAT1 methylation data was obtained from a previous work that also included the patients of CLL#1, using 450k array16. The studied region spanned the genomic coordinates, chr11:65265233–65273940 (hg19) and included 9 CpGs (one in the CpG island, three at the shelf and 5 at the shore) located along the gene body. We also compared the MALAT1 expression in 6 paired samples of CLL and Richter transformation from RNAseq data already published (CLL#2 series, Table 1)38. Finally, we have also analyzed the microarray data from the GSE21029 GEO dataset (CLL#3 series, Table 1), including 17 patients with paired PB/LN samples, and 19 patients with paired PB/BM.
For the analysis of MALAT1 in FL, we extracted RNA from formalin-fixed paraffin-embedded (FFPE) LN samples of 61 FL patients (stages 1 to 3A) from the Hematopathology collection of the Biobank of the Hospital Clínic de Barcelona-IDIBAPS (Spain) (FL#1 series, Table 1). In addition, microarray data generated from sorted FL neoplastic cells of 23 patients were obtained from the GSE107367 GEO dataset40 (FL#2 series, Table 1).
RNA extraction and qRT-PCR
For the 61 FFPE FL samples, 3–10 cuts of 10 µm each were used per sample for RNA extraction using Allprep DNA/RNA FFPE kit (Qiagen). RNA integrity was analyzed with TS4200 using DV200 index as recommended by the manufacturer (Agilent). In this way, samples with DV200 less than 35% were excluded to avoid excessively degraded material. Reverse transcriptase reaction was performed using High-Capacity Reverse transcription kit (Applied Biosystems) with an RNA input of 90 ng.
MALAT1 expression was analyzed by qRT-PCR using a short amplicon specifically designed for FFPE samples. MALAT1 expression levels were normalized using three reference genes (ACTB, GAPDH and YWHAZ) which have been previously used in B-cell lymphomas, including FL samples66,67. Primer quantities were optimized for each amplicon to reach a high efficiency (range: 90–96%) and their sequences were: 5′-CCCCTTCCCTAGGGGATTTCA-3′ (MALAT1 forward), 5′- AAGCCCACAGGAACAAGTCC-3′ (MALAT1 reverse), 5′- CCAACCGCGAGAAGATGAC-3′(ACTB forward), 5′-TAGCACAGCCTGGATAGCAA-3′ (ACTB reverse), 5′- AGGTGAAGGTCGGAGTCA-3′(GAPDH forward), 5′ CAACAATATCCACTTTACCAGAGTTAA-3′ (GAPDH reverse), 5′-CAAAGACAGCACGCTAATAATGCA-3′ (YWHAZ forward), and 5′-TCAGCTTCGTCTCCTTGGGTA-3′ (YWHAZ reverse). Amplification reactions were carried out using PowerUpTm SYBR™ Green Master Mix (Applied Biosystems, Foster City, CA) following supplier’s recommendations in a Step One Plus thermocycler (Applied Biosystems). Transcript expression relative quantification was performed referred to a calibrator sample of universal human reference RNA (Invitrogen).
Bioinformatic analyses and statistics
Microarray data were normalized with RMA-based methodology and used to analyze the expression levels of MALAT1 transcript. Although the expression microarrays were enriched in probe sets for coding genes, several probe sets that hybridize exclusively to the MALAT1 transcript were initially included. The specificity of the probe sets was confirmed in the Affymetrix database (NettAffx™). Only one probe set was excluded (223579_s_at) at this stage. The correlations among the different probe sets were checked by strand and another probe set was excluded as an outlier (224559_at). Finally, the mean values of the remaining probe sets were retained as reliable for measuring the expression levels of MALAT1 (probe sets 1558678_s_at, 223940_x_at, 224558_s_at, 224567_x_at, 224568_x_at and 226675_s_at).
In the clinical association studies, optimal cut-off points for MALAT1 expression groups were obtained using the maxstat algorithm which optimized the log-rank statistics (maxstat package, R Software, Vienna). Cumulative incidence and Kaplan–Meier curves, scatterplots, and box plot graphs were generated using both the R environment and GraphPad Prism v7. Univariate and multivariate Fine-Gray regression models considering MALAT1 as a continuous variable (as a more stringent statistical approach) were used for measuring its impact on time to treatment (TTT). Univariate analysis of MALAT1 as a continuous variable was performed with the Cox regression model for overall survival (OS). Both calculations were performed with R v.4.0.3. Student’s t-test was used to evaluate the differences in the mean expression of MALAT1 among subtypes and molecular factors with previously described prognostic value in CLL. Paired t-test was used to evaluate differences in MALAT1 expression between paired samples of CLL in PB and LN39. Linear regression on scatter plots was representing the correlation between MALAT1 expression and the epiCMIT score (epigenetically-determined Cumulative MIToses)16.
To identify potential target genes, ranked lists of coding genes according to the degree of Pearson’s correlation to MALAT1 were obtained for the different indolent B-cell lymphomas under study using either RNA-seq (CLL#1) or microarray data (FL#2). Only those statistically significant correlations after correction for multiple comparisons (adjusted p-value < 0.05) were considered for the downstream analyses. We separately analyzed positive and negative correlations of coding genes with MALAT1 using the webtool Metascape68. The Metascape output of enriched (Reactome) pathways were obtained as tables, and summary plots depicting relevant pathways with very high statistical significance (q < 0.05) were prepared with Graphpad Prism v7. Furthermore, we used Gene Set Enrichment Analysis (GSEA v3.0 from http://www.gsea-msigdb.org) to perform additional pathway enrichment analyses. First we analyzed the ranked list of coding genes correlated with MALAT1 for M-CLL and U-CLL subtypes using the C2 curated gene signatures from MSigDB/GSEA website and several specific signatures previously related to differences between LN and PB in CLL17. Second, GSEA were applied to global expression matrices for each CLL subtype ranked by their degree of differential expression.
The differential expression analysis comparing samples subsets of MALAT1 high and low expression was performed using Limma in R v.4.0.3. Those subsets were defined using the clinically significant cutoff found in the CLL#1 series, whereas for the FL#2 series the median value of MALAT1 expression was used. The adjustment of p-values for multiple comparisons was performed only in CLL#1, accordingly to the limited sample size of the FL#2 studied series. MORPHEUS software platform (https://software.broadinstitute.org/morpheus) was used to generate the plots using euclidean metrics of hierarchical clusters to depict the significantly overexpressed/downregulated genes found in all these comparisons.
Ethics approval and consent to participate
Samples from FL patients were obtained from the Hematopathology collection of the Biobank of the Hospital Clínic de Barcelona-IDIBAPS (Spain). RNAseq data and full clinical annotations from CLL patients were obtained from the Spanish ICGC consortium, which had an approval of the Institutional Review Board of Hospital Clinic de Barcelona, in accordance with national regulations and the Declaration of Helsinki. All patients provided written informed consent. The remaining data from other samples were obtained from GEO microarray public repository including previously published works with their particular ethical compliances stated in the original articles.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
-
Huang, Y. et al. Regulatory long non-coding RNA and its functions. J. Physiol. Biochem. 68, 611–618 (2012).
Google Scholar
-
Bhan, A., Soleimani, M. & Mandal, S. S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 77, 3965–3981 (2017).
Google Scholar
-
Li, Z.-X. et al. MALAT1: A potential biomarker in cancer. Cancer Manag. Res. 10, 6757–6768 (2018).
Google Scholar
-
Zhang, X., Hamblin, M. H. & Yin, K.-J. The long noncoding RNA Malat 1: Its physiological and pathophysiological functions. RNA Biol. 14, 1705–1714 (2017).
Google Scholar
-
Wang, Y. et al. The long noncoding RNA MALAT-1 is a novel biomarker in various cancers: A meta-analysis based on the GEO database and literature. J. Cancer 7, 991–1001 (2016).
Google Scholar
-
Kwok, Z. H., Roche, V., Chew, X. H., Fadieieva, A. & Tay, Y. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer 143, 668–678 (2018).
Google Scholar
-
Kim, J. et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715 (2018).
Google Scholar
-
Zhou, M. et al. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 16, 16 (2017).
Google Scholar
-
Roisman, A. et al. Differential expression of long non-coding RNAs are related to proliferation and histological diversity in follicular lymphomas. Br. J. Haematol. 184, 373–383 (2019).
Google Scholar
-
Wang, Q.-M., Lian, G.-Y., Song, Y., Huang, Y.-F. & Gong, Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 231, 116335 (2019).
Google Scholar
-
Ahmadi, A. et al. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood Res. 53, 320–324 (2018).
Google Scholar
-
Wang, X. et al. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 14, 346 (2016).
Google Scholar
-
Puente, X. S. et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 526, 519–524 (2015).
Google Scholar
-
Quesada, V. et al. The genomic landscape of chronic lymphocytic leukemia: Clinical implications. BMC Med. 11, 124 (2013).
Google Scholar
-
Nadeu, F. et al. IGLV3-21R110 identifies an aggressive biological subtype of chronic lymphocytic leukemia with intermediate epigenetics. Blood 137, 2935–2946 (2020).
Google Scholar
-
Duran-Ferrer, M. et al. The proliferative history shapes the DNA methylome of B-cell tumors and predicts clinical outcome. Nat. Cancer 1, 1066–1081 (2020).
Google Scholar
-
Dadashian, E. L. et al. TLR signaling is activated in lymph node-resident CLL cells and is only partially inhibited by ibrutinib. Cancer Res 79, 360–371 (2019).
Google Scholar
-
Mittal, A. K. et al. Chronic lymphocytic leukemia cells in a lymph node microenvironment depict molecular signature associated with an aggressive disease. Mol. Med. 20, 290–301 (2014).
Google Scholar
-
Kowalska, W. & Bojarska-Junak, A. Monocytic MDSC as a source of immunosuppressive cytokines in chronic lymphocytic leukemia (CLL) microenvironment. Folia Histochem. Cytobiol. 58, 25–36 (2020).
Google Scholar
-
Mirabilii, S. et al. Biological aspects of mTOR in leukemia. Int. J. Mol. Sci. 19, 2396 (2018).
Google Scholar
-
Lezina, L. et al. CD40L/IL-4-stimulated CLL demonstrates variation in translational regulation of DNA damage response genes including ATM. Blood Adv. 2, 1869–1881 (2018).
Google Scholar
-
Shukla, A., Shukla, V. & Joshi, S. S. Regulation of MAPK signaling and implications in chronic lymphocytic leukemia. Leuk. Lymphoma 59, 1565–1573 (2018).
Google Scholar
-
Zhang, N., Coutinho, L. E. & Pati, D. PDS5A and PDS5B in cohesin function and human disease. Int. J. Mol. Sci. 22, 5868 (2021).
Google Scholar
-
Moon, H. M. et al. LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex. Hum. Mol. Genet. 23, 449–466 (2014).
Google Scholar
-
Lo, F.-Y., Chen, H.-T., Cheng, H.-C., Hsu, H.-S. & Wang, Y.-C. Overexpression of PAFAH1B1 is associated with tumor metastasis and poor survival in non-small cell lung cancer. Lung Cancer 77, 585–592 (2012).
Google Scholar
-
Jiang, Y. et al. NFAT1-Mediated regulation of NDEL1 promotes growth and invasion of glioma stem-like cells. Cancer Res. 79, 2593–2603 (2019).
Google Scholar
-
Hartmann, E. M., Rudelius, M., Burger, J. A. & Rosenwald, A. CCL3 chemokine expression by chronic lymphocytic leukemia cells orchestrates the composition of the microenvironment in lymph node infiltrates. Leuk. Lymphoma 57, 563–571 (2016).
Google Scholar
-
Secchiero, P. et al. Potential pathogenetic implications of cyclooxygenase-2 overexpression in B chronic lymphoid leukemia cells. Am. J. Pathol. 167, 1599–1607 (2005).
Google Scholar
-
Lo, H.-W., Cao, X., Zhu, H. & Ali-Osman, F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol. Cancer Res. 8, 232–245 (2010).
Google Scholar
-
Carpenter, R. L. & Lo, H.-W. STAT3 target genes relevant to human cancers. Cancers (Basel) 6, 897–925 (2014).
Google Scholar
-
Wee, S. et al. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. U. S. A. 105, 13057–13062 (2008).
Google Scholar
-
Mavrakis, K. J. et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 22, 2178–2188 (2008).
Google Scholar
-
Hu, M., Liu, L. & Yao, W. Activation of p53 by costunolide blocks glutaminolysis and inhibits proliferation in human colorectal cancer cells. Gene 678, 261–269 (2018).
Google Scholar
-
Cai, Q. et al. MAPK6-AKT signaling promotes tumor growth and resistance to mTOR kinase blockade. Sci. Adv. 7, eabi6439 (2021).
Google Scholar
-
Horváth, A., Rona, G., Pagano, M. & Jordan, P. W. Interaction between NSMCE4A and GPS1 links the SMC5/6 complex to the COP9 signalosome. BMC Mol. Cell Biol. 21, 36 (2020).
Google Scholar
-
Wang, Z., Potter, C. S., Sundberg, J. P. & Hogenesch, H. SHARPIN is a key regulator of immune and inflammatory responses. J. Cell Mol. Med. 16, 2271–2279 (2012).
Google Scholar
-
Mantovani, F. et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 14, 912–920 (2007).
Google Scholar
-
Nadeu, F. et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat. Med. 28, 1662–1671 (2022).
Google Scholar
-
Herishanu, Y. et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood 117, 563–574 (2011).
Google Scholar
-
Pangault, C. et al. Follicular lymphoma cell niche: Identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia 24, 2080–2089 (2010).
Google Scholar
-
Devan, J., Janikova, A. & Mraz, M. New concepts in follicular lymphoma biology: From BCL2 to epigenetic regulators and non-coding RNAs. Semin. Oncol. 45, 291–302 (2018).
Google Scholar
-
Duś-Szachniewicz, K., Rymkiewicz, G., Agrawal, A. K., Kołodziej, P. & Wiśniewski, J. R. Large-scale proteomic analysis of follicular lymphoma reveals extensive remodeling of cell adhesion pathway and identifies hub proteins related to the lymphomagenesis. Cancers (Basel) 13, 630 (2021).
Google Scholar
-
Dong, T. et al. The expression of CD9 and PIK3CD is associated with prognosis of follicular lymphoma. J. Cancer 6, 1222–1229 (2015).
Google Scholar
-
Saijo, K. et al. Essential role of Src-family protein tyrosine kinases in NF-kappaB activation during B cell development. Nat. Immunol. 4, 274–279 (2003).
Google Scholar
-
Seda, V. & Mraz, M. B-cell receptor signalling and its crosstalk with other pathways in normal and malignant cells. Eur. J. Haematol. 94, 193–205 (2015).
Google Scholar
-
Lee, Y.-J. et al. Prognostic value of CAPZA1 overexpression in gastric cancer. Int. J. Oncol. 42, 1569–1577 (2013).
Google Scholar
-
Qin, T. et al. NcRNA-regulated CAPZA1 associated with prognostic and immunological effects across lung adenocarcinoma. Front. Oncol. 12, 1025192 (2022).
Google Scholar
-
Fu, F., Wei, X., Liu, J. & Mi, N. Bioinformatic analysis of specific genes in diabetic nephropathy. Ren. Fail. 37, 1219–1224 (2015).
Google Scholar
-
Zuehlke, A. D., Beebe, K., Neckers, L. & Prince, T. Regulation and function of the human HSP90AA1 gene. Gene 570, 8–16 (2015).
Google Scholar
-
Pangault, C. et al. Integrative analysis of cell crosstalk within follicular lymphoma cell niche: Towards a definition of the FL supportive synapse. Cancers (Basel) 12, 2865 (2020).
Google Scholar
-
Harjunpää, A. et al. Differential gene expression in non-malignant tumour microenvironment is associated with outcome in follicular lymphoma patients treated with rituximab and CHOP. Br. J. Haematol. 135, 33–42 (2006).
Google Scholar
-
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Google Scholar
-
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).
Google Scholar
-
Guo, F., Guo, L., Li, Y., Zhou, Q. & Li, Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int. J. Clin. Exp. Pathol. 8, 15903–15910 (2015).
Google Scholar
-
Wang, Y. et al. TGF-β-induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR-30a interactions. Cancer Lett. 436, 52–62 (2018).
Google Scholar
-
Zheng, T., Ma, G., Tang, M., Li, Z. & Xu, R. IL-8 secreted from M2 macrophages promoted prostate tumorigenesis via STAT3/MALAT1 pathway. Int. J. Mol. Sci. 20, 98 (2018).
Google Scholar
-
Hong, C.-H., Lin, S.-H. & Lee, C.-H. CCL21 induces mTOR-dependent MALAT1 expression, leading to cell migration in cutaneous T-cell lymphoma. In Vivo 33, 793–800 (2019).
Google Scholar
-
Zhuang, Y.-T. et al. IL-6 induced lncRNA MALAT1 enhances TNF-α expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur. Rev. Med. Pharmacol. Sci. 21, 302–309 (2017).
Google Scholar
-
Stevenson, F. K., Forconi, F. & Kipps, T. J. Exploring the pathways to chronic lymphocytic leukemia. Blood 138, 827–835 (2021).
Google Scholar
-
Ten Hacken, E. & Burger, J. A. Microenvironment interactions and B-cell receptor signaling in chronic lymphocytic leukemia: Implications for disease pathogenesis and treatment. Biochim. Biophys. Acta 1863, 401–413 (2016).
Google Scholar
-
Parry, E. M. et al. Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nat. Med. 29, 158–169 (2023).
Google Scholar
-
Arun, G., Aggarwal, D. & Spector, D. L. MALAT1 long non-coding RNA: Functional implications. Noncoding RNA 6, 22 (2020).
Google Scholar
-
Tauzin, S., Ding, H., Burdevet, D., Borisch, B. & Hoessli, D. C. Membrane-associated signaling in human B-lymphoma lines. Exp. Cell Res. 317, 151–162 (2011).
Google Scholar
-
Ferreira, P. G. et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 24, 212–226 (2014).
Google Scholar
-
Kulis, M. et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 44, 1236–1242 (2012).
Google Scholar
-
Potashnikova, D., Gladkikh, A. & Vorobjev, I. A. Selection of superior reference genes’ combination for quantitative real-time PCR in B-cell lymphomas. Ann. Clin. Lab. Sci. 45, 64–72 (2015).
Google Scholar
-
Yagi, K. et al. Expression of multidrug resistance 1 gene in B-cell lymphomas: association with follicular dendritic cells. Histopathology 62, 414–420 (2013).
Google Scholar
-
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Google Scholar
Acknowledgements
This research was funded by Fundació La Marato de TV3 (“projecte finançat per Fundació La Marató de TV3”) 201920-30 to LH; Ministry of Science and Innovation (MCIN)/AEI(10.13039/501100011033) and European Regional Development Fund “Una manera de hacer Europa” (PID2021-123054OB-I00 to E.C) and the Generalitat de Catalunya Suport Grups de Recerca AGAUR (2021-SGR-01172 to E.C. and 2021-SGR-01293 to S.B.). AGAUR 2018 FIB00696, Generalitat de Catalunya to EMF-G. EC is Academia Researchers of the “Institució Catalana de Recerca i Estudis Avançats” (ICREA) of the Generalitat de Catalunya. We are indebted to the Genomics core facility of the Institut d’Investigacions Biomèdiques August Pi I Sunyer (IDIBAPS).
Author information
Authors and Affiliations
Contributions
Conceived and designed the Study: L.H. and E.C. Collected and processed clinical/expression data from ICGC, Hospital Clinic de Barcelona and GEO: E.M.F.-G., F.N., P.M., A.R., J.D., E.G., A.L.-G., M.D.-F., I.S., C.L., S.B., S.D., P.J., X.S.P. and L.H.; Processed FFPE material from FL samples for RNA purification: S.M. Analyzed the RNAseq/microarrays/qRT-PCR data and performed the statistical analysis: E.M.F.-G., F.N., P.M., A.R. and L.H. Wrote the manuscript: E.M.F.-G., L.H., E.C. and J.I.M.-S. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Table S1.
Supplementary Table S2.
Supplementary Table S3.
Supplementary Table S4.
Supplementary Table S5.
Supplementary Table S6.
Supplementary Table S7.
Supplementary Table S8.
Supplementary Table S9.
Supplementary Table S10.
Supplementary Table S11.
Supplementary Table S12.
Supplementary Table S13.
Supplementary Table S14.
Supplementary Table S15.
Supplementary Table S16.
Supplementary Table S17.
Supplementary Table S18.
Supplementary Figures.
Supplementary Legends.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Fernández-Garnacho, E.M., Nadeu, F., Martín, S. et al. MALAT1 expression is associated with aggressive behavior in indolent B-cell neoplasms.
Sci Rep 13, 16839 (2023). https://doi.org/10.1038/s41598-023-44174-8
-
Received: 15 November 2022
-
Accepted: 04 October 2023
-
Published: 06 October 2023
-
DOI: https://doi.org/10.1038/s41598-023-44174-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.