Infection
Mpox virus infection and drug treatment modelled in human skin organoids
Abstract
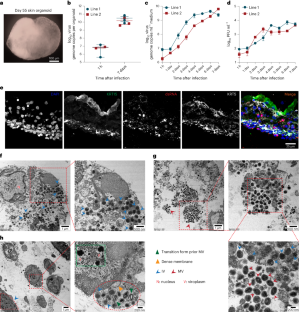
Mpox virus (MPXV) primarily infects human skin to cause lesions. Currently, robust models that recapitulate skin infection by MPXV are lacking. Here we demonstrate that human induced pluripotent stem cell-derived skin organoids are susceptible to MPXV infection and support infectious virus production. Keratinocytes, the predominant cell type of the skin epithelium, effectively support MPXV infection. Using transmission electron microscopy, we visualized the four stages of intracellular virus particle assembly: crescent formation, immature virions, mature virions and wrapped virions. Transcriptional analysis showed that MPXV infection rewires the host transcriptome and triggers abundant expression of viral transcripts. Early treatment with the antiviral drug tecovirimat effectively inhibits infectious virus production and prevents host transcriptome rewiring. Delayed treatment with tecovirimat also inhibits infectious MPXV particle production, albeit to a lesser extent. This study establishes human skin organoids as a robust experimental model for studying MPXV infection, mapping virus–host interactions and testing therapeutics.
This is a preview of subscription content, access via your institution
Access options
style{display:none!important}.LiveAreaSection-193358632 *{align-content:stretch;align-items:stretch;align-self:auto;animation-delay:0s;animation-direction:normal;animation-duration:0s;animation-fill-mode:none;animation-iteration-count:1;animation-name:none;animation-play-state:running;animation-timing-function:ease;azimuth:center;backface-visibility:visible;background-attachment:scroll;background-blend-mode:normal;background-clip:borderBox;background-color:transparent;background-image:none;background-origin:paddingBox;background-position:0 0;background-repeat:repeat;background-size:auto auto;block-size:auto;border-block-end-color:currentcolor;border-block-end-style:none;border-block-end-width:medium;border-block-start-color:currentcolor;border-block-start-style:none;border-block-start-width:medium;border-bottom-color:currentcolor;border-bottom-left-radius:0;border-bottom-right-radius:0;border-bottom-style:none;border-bottom-width:medium;border-collapse:separate;border-image-outset:0s;border-image-repeat:stretch;border-image-slice:100%;border-image-source:none;border-image-width:1;border-inline-end-color:currentcolor;border-inline-end-style:none;border-inline-end-width:medium;border-inline-start-color:currentcolor;border-inline-start-style:none;border-inline-start-width:medium;border-left-color:currentcolor;border-left-style:none;border-left-width:medium;border-right-color:currentcolor;border-right-style:none;border-right-width:medium;border-spacing:0;border-top-color:currentcolor;border-top-left-radius:0;border-top-right-radius:0;border-top-style:none;border-top-width:medium;bottom:auto;box-decoration-break:slice;box-shadow:none;box-sizing:border-box;break-after:auto;break-before:auto;break-inside:auto;caption-side:top;caret-color:auto;clear:none;clip:auto;clip-path:none;color:initial;column-count:auto;column-fill:balance;column-gap:normal;column-rule-color:currentcolor;column-rule-style:none;column-rule-width:medium;column-span:none;column-width:auto;content:normal;counter-increment:none;counter-reset:none;cursor:auto;display:inline;empty-cells:show;filter:none;flex-basis:auto;flex-direction:row;flex-grow:0;flex-shrink:1;flex-wrap:nowrap;float:none;font-family:initial;font-feature-settings:normal;font-kerning:auto;font-language-override:normal;font-size:medium;font-size-adjust:none;font-stretch:normal;font-style:normal;font-synthesis:weight style;font-variant:normal;font-variant-alternates:normal;font-variant-caps:normal;font-variant-east-asian:normal;font-variant-ligatures:normal;font-variant-numeric:normal;font-variant-position:normal;font-weight:400;grid-auto-columns:auto;grid-auto-flow:row;grid-auto-rows:auto;grid-column-end:auto;grid-column-gap:0;grid-column-start:auto;grid-row-end:auto;grid-row-gap:0;grid-row-start:auto;grid-template-areas:none;grid-template-columns:none;grid-template-rows:none;height:auto;hyphens:manual;image-orientation:0deg;image-rendering:auto;image-resolution:1dppx;ime-mode:auto;inline-size:auto;isolation:auto;justify-content:flexStart;left:auto;letter-spacing:normal;line-break:auto;line-height:normal;list-style-image:none;list-style-position:outside;list-style-type:disc;margin-block-end:0;margin-block-start:0;margin-bottom:0;margin-inline-end:0;margin-inline-start:0;margin-left:0;margin-right:0;margin-top:0;mask-clip:borderBox;mask-composite:add;mask-image:none;mask-mode:matchSource;mask-origin:borderBox;mask-position:0 0;mask-repeat:repeat;mask-size:auto;mask-type:luminance;max-height:none;max-width:none;min-block-size:0;min-height:0;min-inline-size:0;min-width:0;mix-blend-mode:normal;object-fit:fill;object-position:50% 50%;offset-block-end:auto;offset-block-start:auto;offset-inline-end:auto;offset-inline-start:auto;opacity:1;order:0;orphans:2;outline-color:initial;outline-offset:0;outline-style:none;outline-width:medium;overflow:visible;overflow-wrap:normal;overflow-x:visible;overflow-y:visible;padding-block-end:0;padding-block-start:0;padding-bottom:0;padding-inline-end:0;padding-inline-start:0;padding-left:0;padding-right:0;padding-top:0;page-break-after:auto;page-break-before:auto;page-break-inside:auto;perspective:none;perspective-origin:50% 50%;pointer-events:auto;position:static;quotes:initial;resize:none;right:auto;ruby-align:spaceAround;ruby-merge:separate;ruby-position:over;scroll-behavior:auto;scroll-snap-coordinate:none;scroll-snap-destination:0 0;scroll-snap-points-x:none;scroll-snap-points-y:none;scroll-snap-type:none;shape-image-threshold:0;shape-margin:0;shape-outside:none;tab-size:8;table-layout:auto;text-align:initial;text-align-last:auto;text-combine-upright:none;text-decoration-color:currentcolor;text-decoration-line:none;text-decoration-style:solid;text-emphasis-color:currentcolor;text-emphasis-position:over right;text-emphasis-style:none;text-indent:0;text-justify:auto;text-orientation:mixed;text-overflow:clip;text-rendering:auto;text-shadow:none;text-transform:none;text-underline-position:auto;top:auto;touch-action:auto;transform:none;transform-box:borderBox;transform-origin:50% 50%0;transform-style:flat;transition-delay:0s;transition-duration:0s;transition-property:all;transition-timing-function:ease;vertical-align:baseline;visibility:visible;white-space:normal;widows:2;width:auto;will-change:auto;word-break:normal;word-spacing:normal;word-wrap:normal;writing-mode:horizontalTb;z-index:auto;-webkit-appearance:none;-moz-appearance:none;-ms-appearance:none;appearance:none;margin:0}.LiveAreaSection-193358632{width:100%}.LiveAreaSection-193358632 .login-option-buybox{display:block;width:100%;font-size:17px;line-height:30px;color:#222;padding-top:30px;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-access-options{display:block;font-weight:700;font-size:17px;line-height:30px;color:#222;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-login>li:not(:first-child)::before{transform:translateY(-50%);content:””;height:1rem;position:absolute;top:50%;left:0;border-left:2px solid #999}.LiveAreaSection-193358632 .additional-login>li:not(:first-child){padding-left:10px}.LiveAreaSection-193358632 .additional-login>li{display:inline-block;position:relative;vertical-align:middle;padding-right:10px}.BuyBoxSection-683559780{display:flex;flex-wrap:wrap;flex:1;flex-direction:row-reverse;margin:-30px -15px 0}.BuyBoxSection-683559780 .box-inner{width:100%;height:100%}.BuyBoxSection-683559780 .readcube-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:1;flex-basis:255px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:300px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox-nature-plus{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:100%;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .title-readcube,.BuyBoxSection-683559780 .title-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .title-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .asia-link{color:#069;cursor:pointer;text-decoration:none;font-size:1.05em;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:1.05em6}.BuyBoxSection-683559780 .access-readcube{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;opacity:.8px;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .price-buybox{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;padding-top:30px;text-align:center}.BuyBoxSection-683559780 .price-buybox-to{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center}.BuyBoxSection-683559780 .price-info-text{font-size:16px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-value{font-size:30px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-per-period{font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-from{font-size:14px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .issue-buybox{display:block;font-size:13px;text-align:center;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:19px}.BuyBoxSection-683559780 .no-price-buybox{display:block;font-size:13px;line-height:18px;text-align:center;padding-right:10%;padding-left:10%;padding-bottom:20px;padding-top:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .vat-buybox{display:block;margin-top:5px;margin-right:20%;margin-left:20%;font-size:11px;color:#222;padding-top:10px;padding-bottom:15px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:17px}.BuyBoxSection-683559780 .tax-buybox{display:block;width:100%;color:#222;padding:20px 16px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:NaNpx}.BuyBoxSection-683559780 .button-container{display:flex;padding-right:20px;padding-left:20px;justify-content:center}.BuyBoxSection-683559780 .button-container>*{flex:1px}.BuyBoxSection-683559780 .button-container>a:hover,.Button-505204839:hover,.Button-1078489254:hover,.Button-2496381730:hover{text-decoration:none}.BuyBoxSection-683559780 .readcube-button{background:#fff;margin-top:30px}.BuyBoxSection-683559780 .button-asia{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;margin-top:75px}.BuyBoxSection-683559780 .button-label-asia,.ButtonLabel-3869432492,.ButtonLabel-3296148077,.ButtonLabel-1651148777{display:block;color:#fff;font-size:17px;line-height:20px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center;text-decoration:none;cursor:pointer}.Button-505204839,.Button-1078489254,.Button-2496381730{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;max-width:320px;margin-top:10px}.Button-505204839 .readcube-label,.Button-1078489254 .readcube-label,.Button-2496381730 .readcube-label{color:#069}
/* style specs end */
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout





Data availability
The data supporting the findings of this study are available within the paper, its supplementary information or its source data. RNA sequencing data are publicly available at https://doi.org/10.17026/dans-xj2-hhat. Mpox viral reads were mapped according to the reference genome (accession NC_063383.1). Source data are provided with this paper.
Code availability
The code used for transcriptomics analysis in this study is publicly available at https://doi.org/10.17026/dans-xzc-46u4
References
-
Gessain, A., Nakoune, E. & Yazdanpanah, Y. Monkeypox. N. Engl. J. Med. 387, 1783–1793 (2022).
Google Scholar
-
Lum, F. M. et al. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat. Rev. Immunol. 22, 597–613 (2022).
Google Scholar
-
Thornhill, J. P. et al. Monkeypox virus infection in humans across 16 Countries—April–June 2022. N. Engl. J. Med. 387, 679–691 (2022).
Google Scholar
-
Zheng, Q. et al. Projecting the impact of testing and vaccination on the transmission dynamics of the 2022 monkeypox outbreak in the USA. J. Travel Med. 29, taac101 (2022).
Google Scholar
-
McCollum, A. M. & Damon, I. K. Human monkeypox. Clin. Infect. Dis. 58, 260–267 (2014).
Google Scholar
-
Adler, H. et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect. Dis. 22, 1153–1162 (2022).
Google Scholar
-
Girometti, N. et al. Demographic and clinical characteristics of confirmed human monkeypox virus cases in individuals attending a sexual health centre in London, UK: an observational analysis. Lancet Infect. Dis. 22, 1321–1328 (2022).
Google Scholar
-
Ogoina, D. et al. Clinical course and outcome of human monkeypox in Nigeria. Clin. Infect. Dis. 71, e210–e214 (2020).
Google Scholar
-
Boesecke, C., Monin, M. B., van Bremen, K., Schlabe, S. & Hoffmann, C. Severe monkeypox-virus infection in undiagnosed advanced HIV infection. Infection 50, 1633–1634 (2022).
Google Scholar
-
Rosa, R. B. et al. In vitro and in vivo models for monkeypox. iScience 26, 105702 (2023).
Google Scholar
-
Lee, J. et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 582, 399–404 (2020).
Google Scholar
-
Jung, S. Y. et al. Wnt-activating human skin organoid model of atopic dermatitis induced by Staphylococcus aureus and its protective effects by Cutibacterium acnes. iScience 25, 105150 (2022).
Google Scholar
-
Ma, J. et al. Establishment of human pluripotent stem cell-derived skin organoids enabled pathophysiological model of SARS-CoV-2 infection. Adv. Sci. 9, e2104192 (2022).
Google Scholar
-
Ma, J. et al. Application of an iPSC-derived organoid model for localized scleroderma therapy. Adv. Sci. 9, e2106075 (2022).
Google Scholar
-
Ramovs, V. et al. Characterization of the epidermal-dermal junction in hiPSC-derived skin organoids. Stem Cell Rep. 17, 1279–1288 (2022).
Google Scholar
-
Lee, J. et al. Generation and characterization of hair-bearing skin organoids from human pluripotent stem cells. Nat. Protoc. 17, 1266–1305 (2022).
Google Scholar
-
Weisberg, A. S. et al. Enigmatic origin of the poxvirus membrane from the endoplasmic reticulum shown by 3D imaging of vaccinia virus assembly mutants. Proc. Natl Acad. Sci. USA 114, E11001–E11009 (2017).
Google Scholar
-
Yang, Z., Bruno, D. P., Martens, C. A., Porcella, S. F. & Moss, B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl Acad. Sci. USA 107, 11513–11518 (2010).
Google Scholar
-
Wilson, S. R. et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell 155, 285–295 (2013).
Google Scholar
-
Lian, L. H., Milora, K. A., Manupipatpong, K. K. & Jensen, L. E. The double-stranded RNA analogue polyinosinic-polycytidylic acid induces keratinocyte pyroptosis and release of IL-36γ. J. Invest. Dermatol. 132, 1346–1353 (2012).
Google Scholar
-
Subramanian, A. et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat. Med. 28, 1706–1714 (2022).
Google Scholar
-
Altindis, M., Puca, E. & Shapo, L. Diagnosis of monkeypox virus—an overview. Travel Med. Infect. Dis. 50, 102459 (2022).
Google Scholar
-
Suner, C., et al. Viral dynamics in patients with monkeypox infection: a prospective cohort study in Spain. Lancet Infect. Dis. 23, 445–453 (2022).
-
Gupta, A. K., Talukder, M., Rosen, T. & Piguet, V. Differential diagnosis, prevention, and treatment of mpox (Monkeypox): a review for dermatologists. Am. J. Clin. Dermatol. 24, 541–556 (2023).
-
Holley, J. et al. Engineered promoter-switched viruses reveal the role of poxvirus maturation protein A26 as a negative regulator of viral spread. J. Virol. 95, e0101221 (2021).
Google Scholar
-
Li, P. et al. Recapitulating infection, thermal sensitivity and antiviral treatment of seasonal coronaviruses in human airway organoids. EBioMedicine 81, 104132 (2022).
Google Scholar
-
Li, P. et al. Recapitulating hepatitis E virus–host interactions and facilitating antiviral drug discovery in human liver-derived organoids. Sci. Adv. 8, eabj5908 (2022).
Google Scholar
-
Yin, Y. et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res 123, 120–131 (2015).
Google Scholar
-
El-Jesr, M., Teir, M. & Maluquer de Motes, C. Vaccinia virus activation and antagonism of cytosolic DNA sensing. Front. Immunol. 11, 568412 (2020).
Google Scholar
-
Albarnaz, J. D. et al. Molecular mimicry of NF-κB by vaccinia virus protein enables selective inhibition of antiviral responses. Nat. Microbiol. 7, 154–168 (2022).
Google Scholar
-
Yang, G. et al. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J. Virol. 79, 13139–13149 (2005).
Google Scholar
-
Grosenbach, D. W. et al. Oral tecovirimat for the treatment of smallpox. N. Engl. J. Med. 379, 44–53 (2018).
Google Scholar
-
Sherwat, A., Brooks, J. T., Birnkrant, D. & Kim, P. Tecovirimat and the treatment of monkeypox—past, present, and future considerations. N. Engl. J. Med. 387, 579–581 (2022).
Google Scholar
-
Desai, A. N. et al. Compassionate use of tecovirimat for the treatment of monkeypox infection. JAMA 328, 1348–1350 (2022).
Google Scholar
-
Frenois-Veyrat, G. et al. Tecovirimat is effective against human monkeypox virus in vitro at nanomolar concentrations. Nat. Microbiol 7, 1951–1955 (2022).
Google Scholar
-
Chinsangaram, J. et al. Pharmacokinetic comparison of a single oral dose of polymorph form i versus form V capsules of the antiorthopoxvirus compound ST-246 in human volunteers. Antimicrob. Agents Chemother. 56, 3582–3586 (2012).
Google Scholar
-
Mondi, A. et al. Clinical experience with use of oral tecovirimat or intravenous cidofovir for the treatment of monkeypox in an Italian reference hospital. J. Infect. 86, 66–117 (2023).
Google Scholar
-
Warner, B. M. et al. In vitro and in vivo efficacy of tecovirimat against a recently emerged 2022 monkeypox virus isolate. Sci. Transl. Med. 14, eade7646 (2022).
Google Scholar
-
O’Laughlin, K. et al. Clinical use of tecovirimat (Tpoxx) for treatment of monkeypox under an investigational new drug protocol—United States, May–August 2022. Morb. Mortal. Wkly Rep. 71, 1190–1195 (2022).
Google Scholar
-
Govind, A. et al. Severe mpox infections in people with uncontrolled human immunodeficiency virus (HIV). Clin. Infect. Dis. 76, 1843–1846 (2023).
Google Scholar
-
McLean, J. et al. Tecovirimat treatment of people with HIV during the 2022 mpox outbreak: a retrospective cohort study. Ann. Intern. Med. 176, 642–648 (2023).
Google Scholar
-
Hutson, C. L. & Damon, I. K. Monkeypox virus infections in small animal models for evaluation of anti-poxvirus agents. Viruses 2, 2763–2776 (2010).
Google Scholar
-
Xuan, D. T. M. et al. Comparison of transcriptomic signatures between monkeypox-infected monkey and human cell lines. J. Immunol. Res 2022, 3883822 (2022).
Google Scholar
-
Zhang, M. et al. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange–Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc. Natl Acad. Sci. USA 111, E5383–E5392 (2014).
Google Scholar
-
Bajanca, F., Luz, M., Duxson, M. J. & Thorsteinsdottir, S. Integrins in the mouse myotome: developmental changes and differences between the epaxial and hypaxial lineage. Dev. Dyn. 231, 402–415 (2004).
Google Scholar
-
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Google Scholar
-
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Google Scholar
-
UCSC Genome Browser on Human (GRCh38/hg38). UCSC https://genome.ucsc.edu/cgi-bin/hgTracks?db=hub_3471181_GCF_014621545.1 (2023).
-
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Google Scholar
Acknowledgements
We thank our colleagues at the hiPSC Hotel, Leiden University Medical Center, for providing hiPS cell lines. This work was supported by a VIDI grant (grant number 91719300) from the Dutch Research Council (NWO) to Q.P., and by the Novo Nordisk Foundation Center for Stem Cell Medicine supported by the Novo Nordisk Foundation, Denmark (grant number NNF21CC0073729) to K.R. K.R. is Chargé de Recherche at the Institut National de la Santé et de la Recherche Médicale (INSERM).
Author information
Authors and Affiliations
Contributions
P.L., S.T.P., K.R. and Q.P. conceptualized the project. G.X., R.S., R.I. and I.A. developed the methodology. P.L., S.T.P., K.R., A.C.V. and Q.P. conducted the investigations. P.L., S.T.P., R.I., I.A. and K.R. conducted formal analysis. P.L., S.T.P., K.R. and Q.P. wrote the original draft. A.C.V., M.J.B. and M.P.P. reviewed and edited the manuscript. K.R. and Q.P. acquired funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks David Ulaeto, Karl Koehler and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quantification of infectious viral titers in organoids at 1 hour and 7 days post-inoculation.
a, Infectious viral titers of organoids after 55 days of differentiation (n = 2 biological replicates). b, Infectious viral titers of organoids after 90 days of differentiation (n = 2 biological replicates).
Source data
Extended Data Fig. 2 Transmission electron microscopy analysis of skin organoids.
(a) and (b) show destroyed organelles, degrading cytoplasm (black arrowheads), and irregular chromatin condensation, alteration of nucleus membrane (red frame) in end-stage skin organoids. c, Representative skin characteristics was visualized in ALI-skin organoid, including skin dermis, collagen fibers, keratin bundles, desmosomes, and basement membrane.
Extended Data Fig. 3 Gene ontology analysis of skin organoids upon MPXV infection.
Top 30 significantly up- and down-regulated pathways by gene ontology analysis at day 7 post-inoculation, compared with the uninfected group. Three independent skin organoids were used in each group, P value determined by DESeq2.
Extended Data Fig. 4 Transcriptomic analysis of skin organoids upon 7 days infection, compared with uninfected or 1 h infection group.
a, Significantly up- and down-regulated genes by volcano plot analysis at day 7 post-inoculation, compared with the uninfected group. P value determined by DESeq2. b-e, Gene set enrichment analysis (GSEA) of KEGG pathways including autophagy_other (HSA04136), apoptosis (HSA04210), necroptosis (HSA04217), and ferroptosis (HSA04216) in skin organoids.
Extended Data Fig. 5 Total reads and mapped MPXV reads in different skin organoid groups by bulk-RNA sequencing.
Skin organoids were either not infected (control), infected for 1 hour (1 hour), infected for 7 days (7 day) or infected and treated with 5 μM of tecovirimat for 7 days (Tecovirimat), n = 3 per group. Data are presented as means of biological replicates ± SEM.
Source data
Extended Data Fig. 6 Low magnification images of the mpox virion (green) and integrin β4 subunit (red) immunostaining depicted in Fig. 6g.
White * indicates the potential autofluorescent signal of the cornified layer of the epithelium, which is distinguishable from the specific staining of mpox virion observed in the conditions where MPXV had been inoculated.
Supplementary information
Supplementary Information
Supplementary table and methods.
Reporting Summary
Source data
Source Data Fig. 1
qPCR and plaque assay source data.
Source Data Fig. 2
qPCR and plaque assay source data.
Source Data Fig. 3
qPCR and plaque assay source data.
Source Data Fig. 5
qPCR and statistical source data.
Source Data Fig. 6
qPCR and statistical source data.
Source Data Extended Data Fig. 1
Plaque assay source data.
Source Data Extended Data Fig. 5
RNA-sequencing mapped reads source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and Permissions
About this article
Cite this article
Li, P., Pachis, S.T., Xu, G. et al. Mpox virus infection and drug treatment modelled in human skin organoids.
Nat Microbiol (2023). https://doi.org/10.1038/s41564-023-01489-6
-
Received: 03 February 2023
-
Accepted: 04 September 2023
-
Published: 12 October 2023
-
DOI: https://doi.org/10.1038/s41564-023-01489-6