Congenital disorders
A fatal case of neonatal viral sepsis caused by human parainfluenza virus type 3
This was a G2P2 (mother: gravida 2, para 2) baby boy with 8 days of age who was born by caesarean section at full term with birth weight 3660 g. His perinatal history was un-eventful. His mother had herpes zoster at 4–5 gestational weeks and common cold at 32 gestational weeks. No family history of genetic diseases was found in his families. Neither the mother nor the infant was found any lab evidence of vertically transmitted diseases, including HIV and CMV. The boy had a healthy elder brother (3 years old).
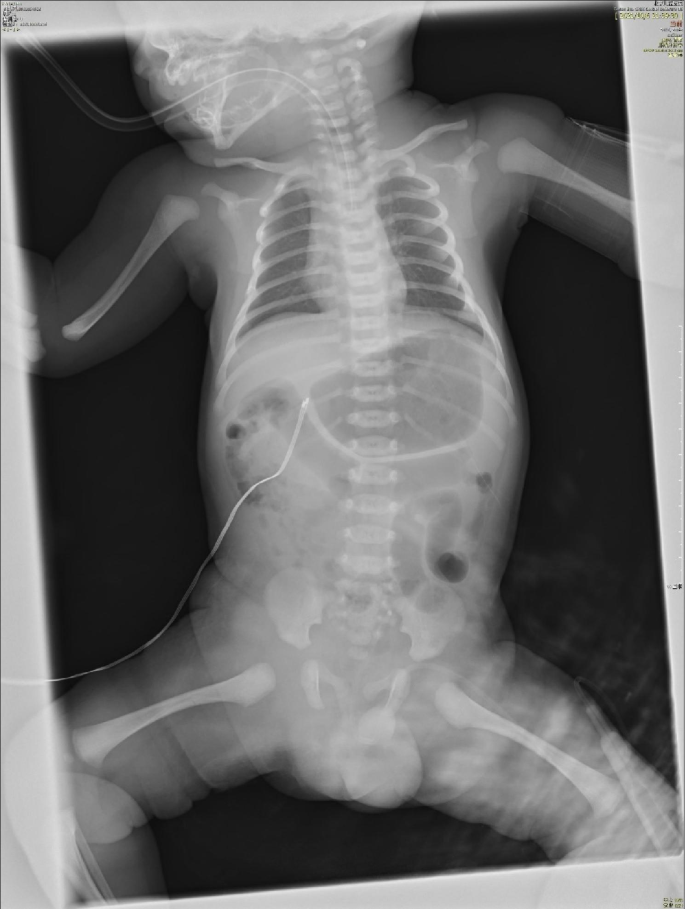
He started to have jaundice on his day of life 4 (DOL 04). The jaundice was slightly progressed but his condition was generally well, and no medical intervention was required. He suddenly presented gross hematuria on DOL 08 with unknown reason. Then he progressively presented gloomy spirit, inactivity and loss of appetite. There was no fever, hematochezia, ecchymosis, or petechiae. After 6 h (17:50, October 06), his parents took him to the Accident & Emergency (A&E) Department of Beijing Children’s Hospital, Capital Medical University, Beijing. He had septic shock when he arrived the A&E Department requiring immediate intubation and mechanical ventilation. Fluid resuscitation and intravenous antibiotics were then started. He was emergently transferred to the neonatal intensive care unit (NICU). His first hemoglobin (Hb) was 122 g/L when he arrived the A&E Department (18:00, October 06), but dropped to 45 g/L in 2 h (Ret 2.57%), requiring repeated packed red blood cell (pRBC) transfusion. His abdominal and chest X-ray found no specific abnormalities (shown in Fig. 1). During his stay in the NICU, he was given 2 times of pRBC (Table 1) due to refractory drop of Hb. No blood loss was found in brain, lungs, abdominal organs and intestine by bed-side ultrasound examination. A urethral catheter was inserted in the A&E Department, but no further urine was collected except for 3ml blood-look urine. His blood gas was severe refractory mixed acidosis. The coagulation test was prothrombin time (PT) 38.4s, fibrinogen (FIB) 0.5 g/L, activated partial prothrombin time (APTT) > 180s(normal range: 11–14 s, 2–4 g/L and 25–37 s, respectively), blood ammonia was 352 µmol/L (normal range: 18–72 µmol/L). He had completely negative response to all anti-shock treatments including fluid resuscitation and vasopressor supports, and died 14 h later.
The anterior-posterior abdominal and chest X-ray in NICU. The first anterior-posterior abdominal and chest X-ray after the patient was admitted to the NICU when he was intubated with low settings of mechanical ventilation. Lungs were normal. Gastric dilatation and slight sausage-like colon were observed. No chest fluid nor abdominal fluid sign was observed. Abbreviation: NICU, neonatal intensive care unit
Five days after death, his blood cultures were reported negative for bacteria, but pharyngeal swab and blood were positive for HPIV-3 virus nucleic acid. Whole exome sequencing did not discover any potential single gene disease or chromosome disorders, such as coagulation abnormalities, congenital metabolic diseases and immunodeficiency diseases. There was no abnormality of copy number variation sequencing (CNV-seq).
Nucleic acid detection by PCR
Nucleic acid testing was performed on pharyngeal swabs and serum samples from the patients. The nucleic acid was extracted using a nucleic acid extraction kit (Da An Gene, Guangzhou, China, Cat. DA0630) and automatic nucleic acid extractor instrument provided by Da An Gene Co., Ltd. The multiplex real-time PCR assay (XABT, Beijing, China) was performed by LightCycler®480 PCR instrument (Roche, Basel, Switzerland). The multiplex PCR kit for 6 respiratory pathogens, including respiratory syncytial virus (RSV), adenovirus (ADV), influenza A virus (Flu A), influenza B virus (Flu B), human parainfluenza virus 1(HPIV-1) and human parainfluenza virus 3(HPIV-3). Both pharyngeal swab and serum sample of the patient showed a positive result in the fluorescent channel of HPIV-3 (Ct values were 17.31 and 35.76, respectively), with CT value less than 36.
Metagenomics next-generation sequencing (mNGS) for pathogen detection
Pharyngeal swabs and serum samples (collected on the day of admission) from the patients were sent to Tianjin Novogene Medical Laboratory for mNGS. The nucleic acid was extracted using QIAamp Viral RNA Mini Kit (52,906, Qiagen Biotech, Germany). RNA libraires were constructed using TIANSeq Fast RNA Library Kit (NR102, Tiangen Biotech, Beijing, China). After the removal of rRNA, RNA fragments of size 150–200 bp were obtained using the enzyme. The first strand cDNA was synthesized, and then the second strand cDNA was synthesized, followed by terminal repair and A-tailing reactions. Adapters were ligated onto the A-tailed fragments. Fragments with adapters were purified and amplified using PCR. After purifying the PCR product, the libraries were pooled and sequenced for 50 bp single ends on an Illumina NovaSeq 6000 machine. The data output of 40 M reads was obtained. Reads were then mapped against the human reference genome using Bowtie2 v2.3.5.1 [11]. After alignment, we removed human reads, the remaining reads were aligned with PD-seq™ database v1.0, which is a self-built database of 9,295 bacteria, 7,210 viruses, 412 fungi and 104 parasites. Kraken2 v2.1.2 [12] and Bracken software v2.6 [13] were used to further analyzed and annotated.
Due to the short time between onset and death, we did not have enough blood samples to detect mNGS, but HIPV-3 was the causative agent of this patient’ death according to the positive result by mNGS in pharyngeal swab and PCR in blood. Additionally, Acinetobacter baumannii (295 reads) and Trichosporon asahii (15 reads) were detected in pharyngeal swab, however bacterial culture of blood specimens was negative, so they are not considered to be the causal pathogens.
Viral molecular analysis
The resultant strain belongs to HPIV-3 and was named HPIV3/BCH/NM (shown in Fig. 2). The whole genome sequence is retrieved by mNGS method and has been deposited in GenBank (accession number: OQ785280). Clean Data were assembled with SOAP denovo software [14, 15]. Phylogenetic analysis of nucleotide sequence based on partial polyprotein gene (HN) or complete genomes of HPIV3/BCH/NM used Neighbor-Joining method with 1000 bootstrap replicates in MEGA 7.0 program. It showed that the HPIV3/BCH/NM strain belongs to Cluster C3a, which is the most common subtype in Beijing. Nucleotide homologies analysis showed that the HPIV3/BCH/NM strain was closely related to the Human respirovirus 3 isolate C256 and HPIV3/06/ZJ/CHN/2017 from China. Comparing with the Wash/47,885/57 prototype strain of HPIV-3, Fourteen amino acid changes were identified in the HN protein region for the strain found in this study. Two unreported amino acid mutations (N86S and S512A) were found in the HN protein region. It is worth noting that S512A lie within a beta − 5 sheet of a key region for the HN protein and cell receptor binding [16].

Phylogenetic analysis of HPIV-3 based on the complete genomes