Congenital disorders
Novel missense variants cause intermediate phenotypes in the phenotypic spectrum of SLC5A6-related disorders
Abstract
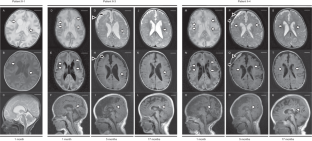
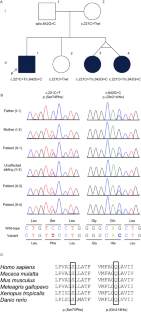
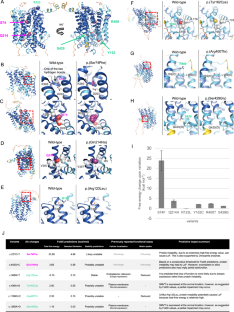
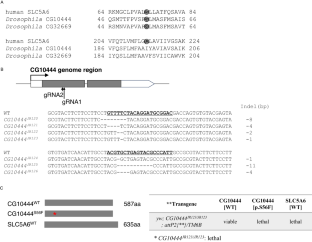
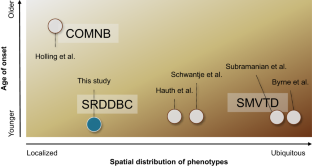
SLC5A6 encodes the sodium-dependent multivitamin transporter, a transmembrane protein that uptakes biotin, pantothenic acid, and lipoic acid. Biallelic SLC5A6 variants cause sodium-dependent multivitamin transporter deficiency (SMVTD) and childhood-onset biotin-responsive peripheral motor neuropathy (COMNB), which both respond well to replacement therapy with the above three nutrients. SMVTD usually presents with various symptoms in multiple organs, such as gastrointestinal hemorrhage, brain atrophy, and global developmental delay, at birth or in infancy. Without nutrient replacement therapy, SMVTD can be lethal in early childhood. COMNB is clinically milder and has a later onset than SMVTD, at approximately 10 years of age. COMNB symptoms are mostly limited to peripheral motor neuropathy. Here we report three patients from one Japanese family harboring novel compound heterozygous missense variants in SLC5A6, namely NM_021095.4:c.[221C>T];[642G>C] p.[(Ser74Phe)];[(Gln214His)]. Both variants were predicted to be deleterious through multiple lines of evidence, including amino acid conservation, in silico predictions of pathogenicity, and protein structure considerations. Drosophila analysis also showed c.221C>T to be pathogenic. All three patients had congenital brain cysts on neonatal cranial imaging, but no other morphological abnormalities. They also had a mild motor developmental delay that almost completely resolved despite no treatment. In terms of severity, their phenotypes were intermediate between SMVTD and COMNB. From these findings we propose a new SLC5A6-related disorder, spontaneously remitting developmental delay with brain cysts (SRDDBC) whose phenotypic severity is between that of SMVTD and COMNB. Further clinical and genetic evidence is needed to support our suggestion.
This is a preview of subscription content, access via your institution
Access options
style{display:none!important}.LiveAreaSection-193358632 *{align-content:stretch;align-items:stretch;align-self:auto;animation-delay:0s;animation-direction:normal;animation-duration:0s;animation-fill-mode:none;animation-iteration-count:1;animation-name:none;animation-play-state:running;animation-timing-function:ease;azimuth:center;backface-visibility:visible;background-attachment:scroll;background-blend-mode:normal;background-clip:borderBox;background-color:transparent;background-image:none;background-origin:paddingBox;background-position:0 0;background-repeat:repeat;background-size:auto auto;block-size:auto;border-block-end-color:currentcolor;border-block-end-style:none;border-block-end-width:medium;border-block-start-color:currentcolor;border-block-start-style:none;border-block-start-width:medium;border-bottom-color:currentcolor;border-bottom-left-radius:0;border-bottom-right-radius:0;border-bottom-style:none;border-bottom-width:medium;border-collapse:separate;border-image-outset:0s;border-image-repeat:stretch;border-image-slice:100%;border-image-source:none;border-image-width:1;border-inline-end-color:currentcolor;border-inline-end-style:none;border-inline-end-width:medium;border-inline-start-color:currentcolor;border-inline-start-style:none;border-inline-start-width:medium;border-left-color:currentcolor;border-left-style:none;border-left-width:medium;border-right-color:currentcolor;border-right-style:none;border-right-width:medium;border-spacing:0;border-top-color:currentcolor;border-top-left-radius:0;border-top-right-radius:0;border-top-style:none;border-top-width:medium;bottom:auto;box-decoration-break:slice;box-shadow:none;box-sizing:border-box;break-after:auto;break-before:auto;break-inside:auto;caption-side:top;caret-color:auto;clear:none;clip:auto;clip-path:none;color:initial;column-count:auto;column-fill:balance;column-gap:normal;column-rule-color:currentcolor;column-rule-style:none;column-rule-width:medium;column-span:none;column-width:auto;content:normal;counter-increment:none;counter-reset:none;cursor:auto;display:inline;empty-cells:show;filter:none;flex-basis:auto;flex-direction:row;flex-grow:0;flex-shrink:1;flex-wrap:nowrap;float:none;font-family:initial;font-feature-settings:normal;font-kerning:auto;font-language-override:normal;font-size:medium;font-size-adjust:none;font-stretch:normal;font-style:normal;font-synthesis:weight style;font-variant:normal;font-variant-alternates:normal;font-variant-caps:normal;font-variant-east-asian:normal;font-variant-ligatures:normal;font-variant-numeric:normal;font-variant-position:normal;font-weight:400;grid-auto-columns:auto;grid-auto-flow:row;grid-auto-rows:auto;grid-column-end:auto;grid-column-gap:0;grid-column-start:auto;grid-row-end:auto;grid-row-gap:0;grid-row-start:auto;grid-template-areas:none;grid-template-columns:none;grid-template-rows:none;height:auto;hyphens:manual;image-orientation:0deg;image-rendering:auto;image-resolution:1dppx;ime-mode:auto;inline-size:auto;isolation:auto;justify-content:flexStart;left:auto;letter-spacing:normal;line-break:auto;line-height:normal;list-style-image:none;list-style-position:outside;list-style-type:disc;margin-block-end:0;margin-block-start:0;margin-bottom:0;margin-inline-end:0;margin-inline-start:0;margin-left:0;margin-right:0;margin-top:0;mask-clip:borderBox;mask-composite:add;mask-image:none;mask-mode:matchSource;mask-origin:borderBox;mask-position:0 0;mask-repeat:repeat;mask-size:auto;mask-type:luminance;max-height:none;max-width:none;min-block-size:0;min-height:0;min-inline-size:0;min-width:0;mix-blend-mode:normal;object-fit:fill;object-position:50% 50%;offset-block-end:auto;offset-block-start:auto;offset-inline-end:auto;offset-inline-start:auto;opacity:1;order:0;orphans:2;outline-color:initial;outline-offset:0;outline-style:none;outline-width:medium;overflow:visible;overflow-wrap:normal;overflow-x:visible;overflow-y:visible;padding-block-end:0;padding-block-start:0;padding-bottom:0;padding-inline-end:0;padding-inline-start:0;padding-left:0;padding-right:0;padding-top:0;page-break-after:auto;page-break-before:auto;page-break-inside:auto;perspective:none;perspective-origin:50% 50%;pointer-events:auto;position:static;quotes:initial;resize:none;right:auto;ruby-align:spaceAround;ruby-merge:separate;ruby-position:over;scroll-behavior:auto;scroll-snap-coordinate:none;scroll-snap-destination:0 0;scroll-snap-points-x:none;scroll-snap-points-y:none;scroll-snap-type:none;shape-image-threshold:0;shape-margin:0;shape-outside:none;tab-size:8;table-layout:auto;text-align:initial;text-align-last:auto;text-combine-upright:none;text-decoration-color:currentcolor;text-decoration-line:none;text-decoration-style:solid;text-emphasis-color:currentcolor;text-emphasis-position:over right;text-emphasis-style:none;text-indent:0;text-justify:auto;text-orientation:mixed;text-overflow:clip;text-rendering:auto;text-shadow:none;text-transform:none;text-underline-position:auto;top:auto;touch-action:auto;transform:none;transform-box:borderBox;transform-origin:50% 50%0;transform-style:flat;transition-delay:0s;transition-duration:0s;transition-property:all;transition-timing-function:ease;vertical-align:baseline;visibility:visible;white-space:normal;widows:2;width:auto;will-change:auto;word-break:normal;word-spacing:normal;word-wrap:normal;writing-mode:horizontalTb;z-index:auto;-webkit-appearance:none;-moz-appearance:none;-ms-appearance:none;appearance:none;margin:0}.LiveAreaSection-193358632{width:100%}.LiveAreaSection-193358632 .login-option-buybox{display:block;width:100%;font-size:17px;line-height:30px;color:#222;padding-top:30px;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-access-options{display:block;font-weight:700;font-size:17px;line-height:30px;color:#222;font-family:Harding,Palatino,serif}.LiveAreaSection-193358632 .additional-login>li:not(:first-child)::before{transform:translateY(-50%);content:””;height:1rem;position:absolute;top:50%;left:0;border-left:2px solid #999}.LiveAreaSection-193358632 .additional-login>li:not(:first-child){padding-left:10px}.LiveAreaSection-193358632 .additional-login>li{display:inline-block;position:relative;vertical-align:middle;padding-right:10px}.BuyBoxSection-683559780{display:flex;flex-wrap:wrap;flex:1;flex-direction:row-reverse;margin:-30px -15px 0}.BuyBoxSection-683559780 .box-inner{width:100%;height:100%}.BuyBoxSection-683559780 .readcube-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:1;flex-basis:255px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:300px;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .subscribe-buybox-nature-plus{background-color:#f3f3f3;flex-shrink:1;flex-grow:4;flex-basis:100%;background-clip:content-box;padding:0 15px;margin-top:30px}.BuyBoxSection-683559780 .title-readcube,.BuyBoxSection-683559780 .title-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .title-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:24px;line-height:32px;color:#222;padding-top:30px;text-align:center;font-family:Harding,Palatino,serif}.BuyBoxSection-683559780 .asia-link{color:#069;cursor:pointer;text-decoration:none;font-size:1.05em;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:1.05em6}.BuyBoxSection-683559780 .access-readcube{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-asia-buybox{display:block;margin:0;margin-right:5%;margin-left:5%;font-size:14px;color:#222;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .access-buybox{display:block;margin:0;margin-right:10%;margin-left:10%;font-size:14px;color:#222;opacity:.8px;padding-top:10px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .price-buybox{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;padding-top:30px;text-align:center}.BuyBoxSection-683559780 .price-buybox-to{display:block;font-size:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center}.BuyBoxSection-683559780 .price-info-text{font-size:16px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-value{font-size:30px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-per-period{font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .price-from{font-size:14px;padding-right:10px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:20px}.BuyBoxSection-683559780 .issue-buybox{display:block;font-size:13px;text-align:center;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:19px}.BuyBoxSection-683559780 .no-price-buybox{display:block;font-size:13px;line-height:18px;text-align:center;padding-right:10%;padding-left:10%;padding-bottom:20px;padding-top:30px;color:#222;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif}.BuyBoxSection-683559780 .vat-buybox{display:block;margin-top:5px;margin-right:20%;margin-left:20%;font-size:11px;color:#222;padding-top:10px;padding-bottom:15px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:17px}.BuyBoxSection-683559780 .tax-buybox{display:block;width:100%;color:#222;padding:20px 16px;text-align:center;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;line-height:NaNpx}.BuyBoxSection-683559780 .button-container{display:flex;padding-right:20px;padding-left:20px;justify-content:center}.BuyBoxSection-683559780 .button-container>*{flex:1px}.BuyBoxSection-683559780 .button-container>a:hover,.Button-1078489254:hover,.Button-2496381730:hover{text-decoration:none}.BuyBoxSection-683559780 .readcube-button{background:#fff;margin-top:30px}.BuyBoxSection-683559780 .button-asia{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;margin-top:75px}.BuyBoxSection-683559780 .button-label-asia,.ButtonLabel-3296148077,.ButtonLabel-1651148777{display:block;color:#fff;font-size:17px;line-height:20px;font-family:-apple-system,BlinkMacSystemFont,”Segoe UI”,Roboto,Oxygen-Sans,Ubuntu,Cantarell,”Helvetica Neue”,sans-serif;text-align:center;text-decoration:none;cursor:pointer}.Button-1078489254,.Button-2496381730{background:#069;border:1px solid #069;border-radius:0;cursor:pointer;display:block;padding:9px;outline:0;text-align:center;text-decoration:none;min-width:80px;max-width:320px;margin-top:10px}.Button-1078489254 .readcube-label,.Button-2496381730 .readcube-label{color:#069}
/* style specs end */
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout

References
-
Wang H, Huang W, Fei Y-J, Xia H, Yang-Feng TL, Leibach FH, et al. Human placental Na+-dependent multivitamin transporter. J Biol Chem. 1999;274:14875–83.
Google Scholar
-
Baumgartner MR, Suormala T. Biotin-responsive Disorders. In: Inborn Metabolic Diseases. Springer Berlin Heidelberg. 2016. p. 375–83.
-
Byrne AB, Arts P, Polyak SW, Feng J, Schreiber AW, Kassahn KS, et al. Identification and targeted management of a neurodegenerative disorder caused by biallelic mutations in SLC5A6. NPJ Genom Med. 2019;14:28.
Google Scholar
-
Hauth I, Waterham H, Wanders RJ, Van Der Crabben S, Van Karnebeek CD. A mild case of SMVT deficiency illustrating the importance of treatment response in variant classification. Mol Case Stud. 2022;8:a006185.
Google Scholar
-
Holling T, Nampoothiri S, Tarhan B, Schneeberger PE, Vinayan KP, Yesodharan D, et al. Novel biallelic variants expand the SLC5A6-related phenotypic spectrum. Eur J Hum Genet. 2022;30:439–49.
Google Scholar
-
Subramanian VS, Constantinescu AR, Benke PJ, Said HM. Mutations in SLC5A6 associated with brain, immune, bone, and intestinal dysfunction in a young child. Hum Genet. 2017;136:253–61.
Google Scholar
-
Schwantje M, De Sain-Van Der Velden M, Jans J, Van Gassen K, Dorrepaal C, Koop K, et al. Genetic defect of the sodium‐dependent multivitamin transporter: a treatable disease, mimicking biotinidase deficiency. JIMD Rep. 2019;48:11–14.
Google Scholar
-
Kameyama S, Mizuguchi T, Fukuda H, Moey LH, Keng WT, Okamoto N, et al. Biallelic null variants in ZNF142 cause global developmental delay with familial epilepsy and dysmorphic features. J Hum Genet. 2022;67:169–73.
Google Scholar
-
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Google Scholar
-
Ng PC. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–14.
Google Scholar
-
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Google Scholar
-
Steinhaus R, Proft S, Schuelke M, Cooper DN, Schwarz Jana M, Seelow D. MutationTaster2021. Nucleic Acids Res. 2021;49:W446–51.
Google Scholar
-
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–15.
Google Scholar
-
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–85.
Google Scholar
-
Capriotti E, Altman RB, Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genom. 2013;14:S2.
Google Scholar
-
Danzi MC, Dohrn MF, Fazal S, Beijer D, Rebelo AP, Cintra V, et al. Deep structured learning for variant prioritization in Mendelian diseases. Nat Commun. 2023;14:4167.
Google Scholar
-
Cheng J, Novati G, Pan J, Bycroft C, Žemgulytė A, Applebaum T, et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science. 2023;381:eadg7492.
Google Scholar
-
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80.
Google Scholar
-
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–89.
Google Scholar
-
Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–87.
Google Scholar
-
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–8.
Google Scholar
-
Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in drosophila. Genetics. 2013;195:715–21.
Google Scholar
-
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Google Scholar
-
Tadaka S, Hishinuma E, Komaki S, Motoike IN, Kawashima J, Saigusa D, et al. jMorp updates in 2020: large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 2021;49:D536–D44.
Google Scholar
-
Gerasimavicius L, Liu X, Marsh JA. Identification of pathogenic missense mutations using protein stability predictors. Sci Rep. 2020;10:15387.
Google Scholar
-
Bromberg Y, Rost B. Correlating protein function and stability through the analysis of single amino acid substitutions. BMC Bioinforma. 2009;10:S8.
Google Scholar
-
Institute of Medicine Standing Committee on the Scientific Evaluation of Dietary Reference I, its Panel on Folate OBV, Choline. The National Academies Collection: Reports funded by National Institutes of Health. In: Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B(6), Folate, Vitamin B(12), Pantothenic Acid, Biotin, and Choline. Washington (DC): National Academies Press (US)Copyright © 1998, National Academy of Sciences.; 1998.
-
Tokutomi T, Fukushima A, Yamamoto K, Bansho Y, Hachiya T, Shimizu A. f-treeGC: a questionnaire-based family tree-creation software for genetic counseling and genome cohort studies. BMC Med Genet. 2017;18:71.
Google Scholar
Acknowledgements
We thank the participants for their cooperation. We also thank Ms. K. Takabe, Mr. T. Miyama, Ms. N. Watanabe, Ms. M. Sato, and Ms. S. Sugimoto at the Department of Human Genetics, Yokohama City University Graduate School of Medicine, for their technical assistance. We also thank Jeremy Allen, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Funding
This work was supported by the Japan Agency for Medical Research and Development (AMED) under grant numbers JP23ek0109674, JP23ek0109549, JP23ek0109617 and JP23ek0109648 (N. Matsumoto); JSPS KAKENHI under grant numbers JP23H02829 (S. Miyatake), JP23K15353 (E. Koshimizu), JP22K15646 (K. Hamanaka), JP22K15901 (A. Fujita), JP23K07229 (Y. Uchiyama), JP23K15353 (N. Tsuchida), and JP23H02877 (T. Mizuguchi); the Takeda Science Foundation (T. Mizuguchi, and N. Matsumoto).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
README
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and Permissions
About this article
Cite this article
Utsuno, Y., Hamada, K., Hamanaka, K. et al. Novel missense variants cause intermediate phenotypes in the phenotypic spectrum of SLC5A6-related disorders.
J Hum Genet (2023). https://doi.org/10.1038/s10038-023-01206-5
-
Received: 23 January 2023
-
Revised: 05 November 2023
-
Accepted: 05 November 2023
-
Published: 27 November 2023
-
DOI: https://doi.org/10.1038/s10038-023-01206-5
